Use of the Microbiome in the Practice of Epidemiology: A Primer on -Omic Technologies
- PMID: 26025238
- PMCID: PMC4498138
- DOI: 10.1093/aje/kwv102
Use of the Microbiome in the Practice of Epidemiology: A Primer on -Omic Technologies
Abstract
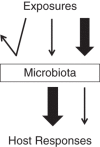
The term microbiome refers to the collective genome of the microbes living in and on our bodies, but it has colloquially come to mean the bacteria, viruses, archaea, and fungi that make up the microbiota (previously known as microflora). We can identify the microbes present in the human body (membership) and their relative abundance using genomics, characterize their genetic potential (or gene pool) using metagenomics, and describe their ongoing functions using transcriptomics, proteomics, and metabolomics. Epidemiologists can make a major contribution to this emerging field by performing well-designed, well-conducted, and appropriately powered studies and by including measures of microbiota in current and future cohort studies to characterize natural variation in microbiota composition and function, identify important confounders and effect modifiers, and generate and test hypotheses about the role of microbiota in health and disease. In this review, we provide an overview of the rapidly growing literature on the microbiome, describe which aspects of the microbiome can be measured and how, and discuss the challenges of including the microbiome as either an exposure or an outcome in epidemiologic studies.
Keywords: bioinformatics; diversity; genomics; microbiome; microbiota.
© The Author 2015. Published by Oxford University Press on behalf of the Johns Hopkins Bloomberg School of Public Health. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

