Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella
- PMID: 26025440
- PMCID: PMC4702336
- DOI: 10.1186/s13059-015-0677-2
Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella
Abstract
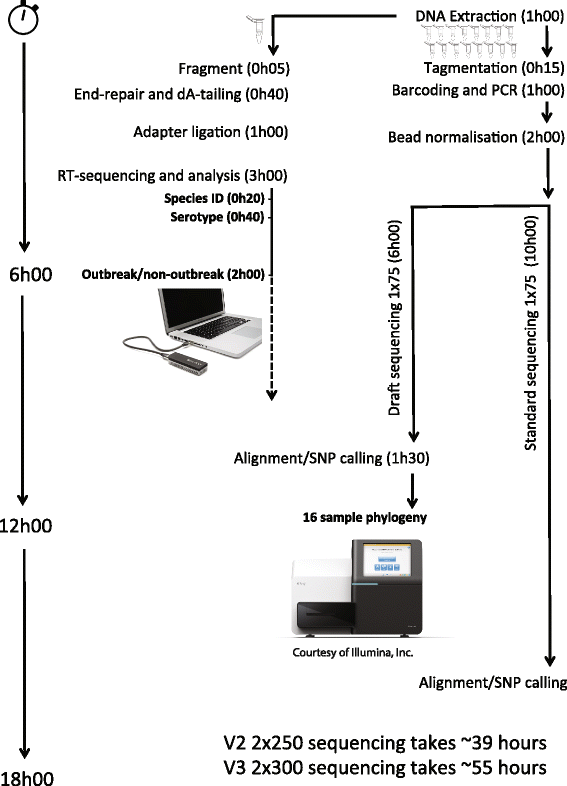
Background: Foodborne outbreaks of Salmonella remain a pressing public health concern. We recently detected a large outbreak of Salmonella enterica serovar Enteritidis phage type 14b affecting more than 30 patients in our hospital. This outbreak was linked to community, national and European-wide cases. Hospital patients with Salmonella are at high risk, and require a rapid response. We initially investigated this outbreak by whole-genome sequencing using a novel rapid protocol on the Illumina MiSeq; we then integrated these data with whole-genome data from surveillance sequencing, thereby placing the outbreak in a national context. Additionally, we investigated the potential of a newly released sequencing technology, the MinION from Oxford Nanopore Technologies, in the management of a hospital outbreak of Salmonella.
Results: We demonstrate that rapid MiSeq sequencing can reduce the time to answer compared to the standard sequencing protocol with no impact on the results. We show, for the first time, that the MinION can acquire clinically relevant information in real time and within minutes of a DNA library being loaded. MinION sequencing permits confident assignment to species level within 20 min. Using a novel streaming phylogenetic placement method samples can be assigned to a serotype in 40 min and determined to be part of the outbreak in less than 2 h.
Conclusions: Both approaches yielded reliable and actionable clinical information on the Salmonella outbreak in less than half a day. The rapid availability of such information may facilitate more informed epidemiological investigations and influence infection control practices.
Figures
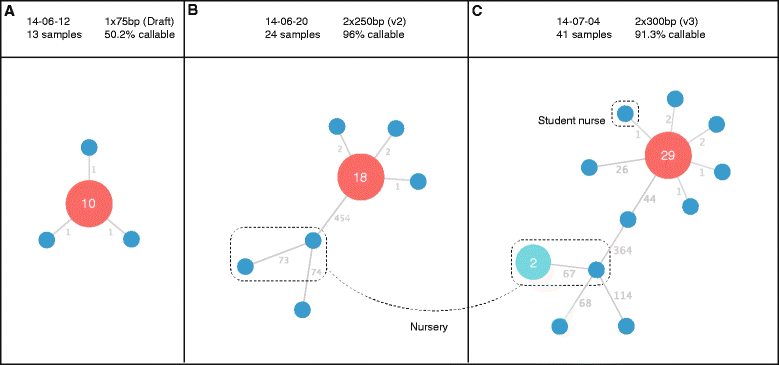
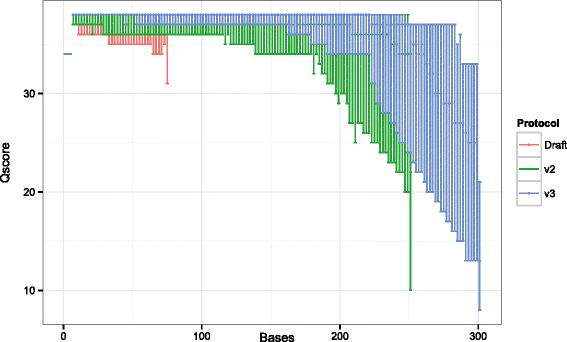

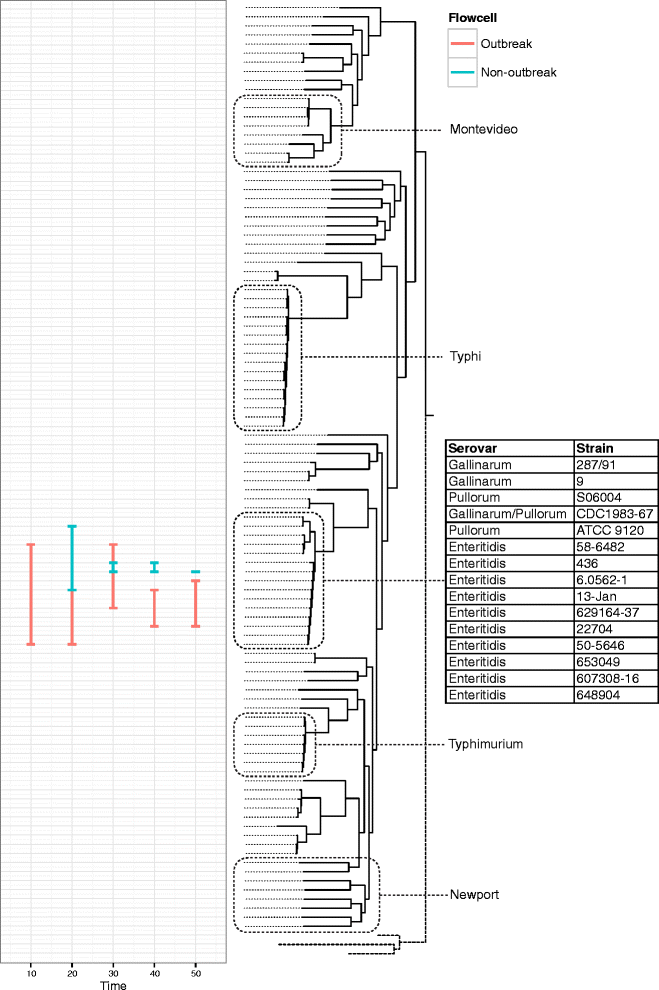
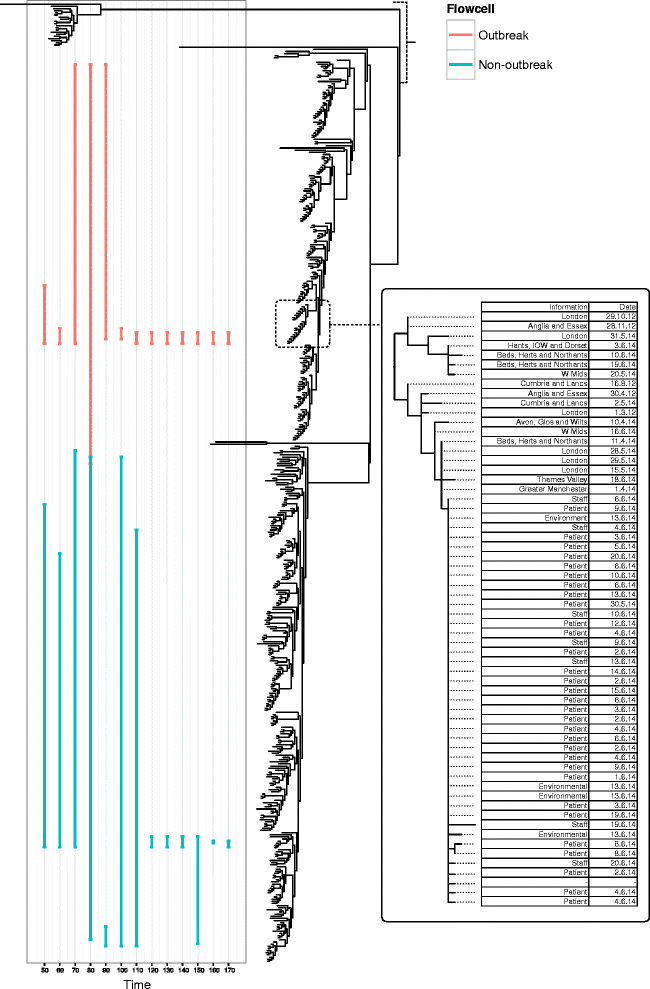
References
-
- Mason BW, Williams N, Salmon RL, Lewis A, Price J, Johnston KM, et al. Outbreak of Salmonella indiana associated with egg mayonnaise sandwiches at an acute NHS hospital. Commun Dis Public Health. 2001;4:300–4. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

