Outwitting EF-Tu and the ribosome: translation with d-amino acids
- PMID: 26026160
- PMCID: PMC4499158
- DOI: 10.1093/nar/gkv566
Outwitting EF-Tu and the ribosome: translation with d-amino acids
Abstract
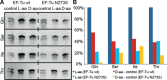
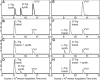
Key components of the translational apparatus, i.e. ribosomes, elongation factor EF-Tu and most aminoacyl-tRNA synthetases, are stereoselective and prevent incorporation of d-amino acids (d-aa) into polypeptides. The rare appearance of d-aa in natural polypeptides arises from post-translational modifications or non-ribosomal synthesis. We introduce an in vitro translation system that enables single incorporation of 17 out of 18 tested d-aa into a polypeptide; incorporation of two or three successive d-aa was also observed in several cases. The system consists of wild-type components and d-aa are introduced via artificially charged, unmodified tRNA(Gly) that was selected according to the rules of 'thermodynamic compensation'. The results reveal an unexpected plasticity of the ribosomal peptidyltransferase center and thus shed new light on the mechanism of chiral discrimination during translation. Furthermore, ribosomal incorporation of d-aa into polypeptides may greatly expand the armamentarium of in vitro translation towards the identification of peptides and proteins with new properties and functions.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Kreil G. D-amino acids in animal peptides. Annu. Rev. Biochem. 1997;66:337–345. - PubMed
-
- Luo L., Kohli R.M., Onishi M., Linne U., Marahiel M.A., Walsh C.T. Timing of epimerization and condensation reactions in nonribosomal peptide assembly lines: kinetic analysis of phenylalanine activating elongation modules of tyrocidine synthetase B. Biochemistry. 2002;41:9184–9196. - PubMed
-
- Chen S., Gfeller D., Buth S.A., Michielin O., Leiman P.G., Heinis C. Improving binding affinity and stability of peptide ligands by substituting glycines with D-amino acids. Chembiochem. 2013;14:1316–1322. - PubMed
-
- Soutourina J., Plateau P., Blanquet S. Metabolism of D-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiae cells. J. Biol. Chem. 2000;275:32535–32542. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

