Analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms
- PMID: 26028266
- PMCID: PMC4451611
- DOI: 10.1038/ncomms8256
Analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms
Erratum in
-
Erratum: analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms.Nat Commun. 2015 Sep 8;6:8389. doi: 10.1038/ncomms9389. Nat Commun. 2015. PMID: 26346554 No abstract available.
Abstract
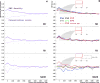
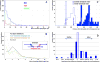
Investigating genomic structural variants at basepair resolution is crucial for understanding their formation mechanisms. We identify and analyse 8,943 deletion breakpoints in 1,092 samples from the 1000 Genomes Project. We find breakpoints have more nearby SNPs and indels than the genomic average, likely a consequence of relaxed selection. By investigating the correlation of breakpoints with DNA methylation, Hi-C interactions, and histone marks and the substitution patterns of nucleotides near them, we find that breakpoints with the signature of non-allelic homologous recombination (NAHR) are associated with open chromatin. We hypothesize that some NAHR deletions occur without DNA replication and cell division, in embryonic and germline cells. In contrast, breakpoints associated with non-homologous (NH) mechanisms often have sequence microinsertions, templated from later replicating genomic sites, spaced at two characteristic distances from the breakpoint. These microinsertions are consistent with template-switching events and suggest a particular spatiotemporal configuration for DNA during the events.
Conflict of interest statement
Authors declare no competing financial interests.
Figures


References
-
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. - PubMed
-
- Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu Rev Genomics Hum Genet. 2006;7:407–442. - PubMed
-
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

