Fibroblast Growth Factor Signaling Controls Liver Size in Mice With Humanized Livers
- PMID: 26028580
- PMCID: PMC4550566
- DOI: 10.1053/j.gastro.2015.05.043
Fibroblast Growth Factor Signaling Controls Liver Size in Mice With Humanized Livers
Abstract
Background & aims: The ratio of liver size to body weight (hepatostat) is tightly controlled, but little is known about how the physiologic functions of the liver help determine its size. Livers of mice repopulated with human hepatocytes (humanized livers) grow to larger than normal; the human hepatocytes do not recognize the fibroblast growth factor (FGF)-15 produced by mouse intestine. This results in up-regulation of bile acid synthesis in the human hepatocytes and enlargement of the bile acid pool. We investigated whether abnormal bile acid signaling affects the hepatostat in mice.
Methods: We crossed Fah(-/-), Rag2(-/-), Il2r(-/-) mice with nonobese diabetic mice to create FRGN mice, whose livers can be fully repopulated with human hepatocytes. We inserted the gene for human FGF19 (ortholog to mouse Fgf15), including regulatory sequences, into the FRGN mice to create FRGN19(+) mice. Livers of FRGN19(+) mice and their FRGN littermates were fully repopulated with human hepatocytes. Liver tissues were collected and bile acid pool sizes and RNA sequences were analyzed and compared with those of mice without humanized livers (controls).
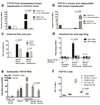
Results: Livers were larger in FRGN mice with humanized livers (13% of body weight), compared with control FRGN mice; they also had much larger bile acid pools and aberrant bile acid signaling. Livers from FRGN19(+) normalized to 7.8% of body weight, and their bile acid pool and signaling more closely resembled that of control FRGN19(+) mice. RNA sequence analysis showed activation of the Hippo pathway, and immunohistochemical and transcription analyses revealed increased hepatocyte proliferation, but not apoptosis, in the enlarged humanized livers of FRGN mice. Cell sorting experiments showed that although healthy human liver does not produce FGF19, nonparenchymal cells from cholestatic livers produce FGF19.
Conclusions: In mice with humanized livers, expression of an FGF19 transgene corrects bile acid signaling defects, resulting in normalization of bile acid synthesis, the bile acid pool, and liver size. These findings indicate that liver size is, in part, regulated by the size of the bile acid pool that the liver must circulate.
Keywords: CYP7A; Mouse Model; Regeneration; Signal Transduction.
Copyright © 2015 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest related to this manuscript.
Figures




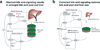
Comment in
-
The FXR-FGF19 Gut-Liver Axis as a Novel "Hepatostat".Gastroenterology. 2015 Sep;149(3):537-40. doi: 10.1053/j.gastro.2015.07.029. Epub 2015 Jul 27. Gastroenterology. 2015. PMID: 26226571 No abstract available.
References
-
- Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. - PubMed
-
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. - PubMed
-
- Camargo FD, Gokhale S, Johnnidis JB, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

