The Min system and other nucleoid-independent regulators of Z ring positioning
- PMID: 26029202
- PMCID: PMC4429545
- DOI: 10.3389/fmicb.2015.00478
The Min system and other nucleoid-independent regulators of Z ring positioning
Abstract
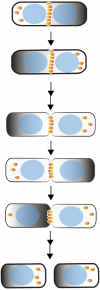
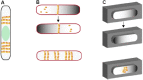
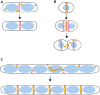
Rod-shaped bacteria such as E. coli have mechanisms to position their cell division plane at the precise center of the cell, to ensure that the daughter cells are equal in size. The two main mechanisms are the Min system and nucleoid occlusion (NO), both of which work by inhibiting assembly of FtsZ, the tubulin-like scaffold that forms the cytokinetic Z ring. Whereas NO prevents Z rings from constricting over unsegregated nucleoids, the Min system is nucleoid-independent and even functions in cells lacking nucleoids and thus NO. The Min proteins of E. coli and B. subtilis form bipolar gradients that inhibit Z ring formation most at the cell poles and least at the nascent division plane. This article will outline the molecular mechanisms behind Min function in E. coli and B. subtilis, and discuss distinct Z ring positioning systems in other bacterial species.
Keywords: FtsZ; Min system; Z-ring positioning; bacterial cell division; divisome.
Figures

References
-
- Åkerlund T., Gullbrand B., Nordström K. (2002). Effects of the Min system on nucleoid segregation in Escherichia coli. Microbiology 148, 3213–3222. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

