Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients
- PMID: 26030179
- PMCID: PMC4868598
- DOI: 10.1038/nm.3870
Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients
Erratum in
- Nat Med. 2015 Jul;21(7):doi:10.1038/nm0715-827b
-
Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients.Nat Med. 2015 Jul;21(7):827. doi: 10.1038/nm0715-827b. Nat Med. 2015. PMID: 26151329 No abstract available.
Abstract
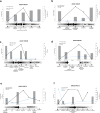
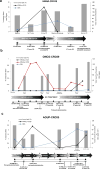
Colorectal cancers (CRCs) evolve by a reiterative process of genetic diversification and clonal evolution. The molecular profile of CRC is routinely assessed in surgical or bioptic samples. Genotyping of CRC tissue has inherent limitations; a tissue sample represents a single snapshot in time, and it is subjected to spatial selection bias owing to tumor heterogeneity. Repeated tissue samples are difficult to obtain and cannot be used for dynamic monitoring of disease progression and response to therapy. We exploited circulating tumor DNA (ctDNA) to genotype colorectal tumors and track clonal evolution during treatment with the epidermal growth factor receptor (EGFR)-specific antibodies cetuximab or panitumumab. We identified alterations in ctDNA of patients with primary or acquired resistance to EGFR blockade in the following genes: KRAS, NRAS, MET, ERBB2, FLT3, EGFR and MAP2K1. Mutated KRAS clones, which emerge in blood during EGFR blockade, decline upon withdrawal of EGFR-specific antibodies, indicating that clonal evolution continues beyond clinical progression. Pharmacogenomic analysis of CRC cells that had acquired resistance to cetuximab reveals that upon antibody withdrawal KRAS clones decay, whereas the population regains drug sensitivity. ctDNA profiles of individuals who benefit from multiple challenges with anti-EGFR antibodies exhibit pulsatile levels of mutant KRAS. These results indicate that the CRC genome adapts dynamically to intermittent drug schedules and provide a molecular explanation for the efficacy of rechallenge therapies based on EGFR blockade.
Figures


Comment in
-
Colorectal cancer: Liquid biopsy enables real-time monitoring of molecular alterations in CRC.Nat Rev Gastroenterol Hepatol. 2015 Jul;12(7):372. doi: 10.1038/nrgastro.2015.105. Epub 2015 Jun 16. Nat Rev Gastroenterol Hepatol. 2015. PMID: 26077554 No abstract available.
-
Gastrointestinal cancer: Tracking tumour evolution through liquid biopsy.Nat Rev Clin Oncol. 2015 Oct;12(10):565-6. doi: 10.1038/nrclinonc.2015.153. Epub 2015 Sep 8. Nat Rev Clin Oncol. 2015. PMID: 26346844 No abstract available.
References
-
- Douillard JY, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. - PubMed
-
- Di Nicolantonio F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. - PubMed
-
- Bertotti A, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

