Genetic basis of autoimmunity
- PMID: 26030227
- PMCID: PMC4497748
- DOI: 10.1172/JCI78086
Genetic basis of autoimmunity
Abstract
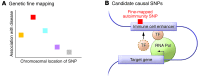
Autoimmune diseases affect up to approximately 10% of the population. While rare Mendelian autoimmunity syndromes can result from monogenic mutations disrupting essential mechanisms of central and peripheral tolerance, more common human autoimmune diseases are complex disorders that arise from the interaction between polygenic risk factors and environmental factors. Although the risk attributable to most individual nucleotide variants is modest, genome-wide association studies (GWAS) have the potential to provide an unbiased view of biological pathways that drive human autoimmune diseases. Interpretation of GWAS requires integration of multiple genomic datasets including dense genotyping, cis-regulatory maps of primary immune cells, and genotyped studies of gene expression in relevant cell types and cellular conditions. Improved understanding of the genetic basis of autoimmunity may lead to a more sophisticated understanding of underlying cellular phenotypes and, eventually, novel diagnostics and targeted therapies.
Figures

References
-
- Sadovnick AD, et al. A population-based study of multiple sclerosis in twins: update. Ann Neurol. 1993;33(3):281–285. - PubMed

