Cancer Cells Hijack PRC2 to Modify Multiple Cytokine Pathways
- PMID: 26030458
- PMCID: PMC4450877
- DOI: 10.1371/journal.pone.0126466
Cancer Cells Hijack PRC2 to Modify Multiple Cytokine Pathways
Abstract
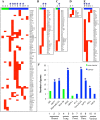
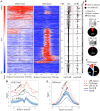
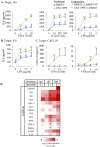
Polycomb Repressive Complex 2 (PRC2) is an epigenetic regulator induced in many cancers. It is thought to drive tumorigenesis by repressing division, stemness, and/or developmental regulators. Cancers evade immune detection, and diverse immune regulators are perturbed in different tumors. It is unclear how such cell-specific effects are coordinated. Here, we show a profound and cancer-selective role for PRC2 in repressing multiple cytokine pathways. We find that PRC2 represses hundreds of IFNγ stimulated genes (ISGs), cytokines and cytokine receptors. This target repertoire is significantly broadened in cancer vs non-cancer cells, and is distinct in different cancer types. PRC2 is therefore a higher order regulator of the immune program in cancer cells. Inhibiting PRC2 with either RNAi or EZH2 inhibitors activates cytokine/cytokine receptor promoters marked with bivalent H3K27me3/H3K4me3 chromatin, and augments responsiveness to diverse immune signals. PRC2 inhibition rescues immune gene induction even in the absence of SWI/SNF, a tumor suppressor defective in ~20% of human cancers. This novel PRC2 function in tumor cells could profoundly impact the mechanism of action and efficacy of EZH2 inhibitors in cancer treatment.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

