Genome-wide mining, characterization, and development of microsatellite markers in gossypium species
- PMID: 26030481
- PMCID: PMC4650602
- DOI: 10.1038/srep10638
Genome-wide mining, characterization, and development of microsatellite markers in gossypium species
Abstract
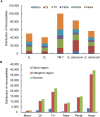
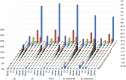
Although much research has been conducted to characterize microsatellites and develop markers, the distribution of microsatellites remains ambiguous and the use of microsatellite markers in genomic studies and marker-assisted selection is limited. To identify microsatellites for cotton research, we mined 100,290, 83,160, and 56,937 microsatellites with frequencies of 41.2, 49.1, and 74.8 microsatellites per Mb in the recently sequenced Gossypium species: G. hirsutum, G. arboreum, and G. raimondii, respectively. The distributions of microsatellites in their genomes were non-random and were positively and negatively correlated with genes and transposable elements, respectively. Of the 77,996 developed microsatellite markers, 65,498 were physically anchored to the 26 chromosomes of G. hirsutum with an average marker density of 34 markers per Mb. We confirmed 67,880 (87%) universal and 7,705 (9.9%) new genic microsatellite markers. The polymorphism was estimated in above three species by in silico PCR and validated with 505 markers in G. hirsutum. We further predicted 8,825 polymorphic microsatellite markers within G. hirsutum acc. TM-1 and G. barbadense cv. Hai7124. In our study, genome-wide mining and characterization of microsatellites, and marker development were very useful for the saturation of the allotetraploid genetic linkage map, genome evolution studies and comparative genome mapping.
Figures
References
-
- Gupta P. & Varshney R. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113, 163–185 (2000).
-
- Powell W., Machray G. & Provan J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1, 215–222 (1996).
-
- Agarwal M., Shrivastava N. & Padh H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 27, 617–631 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

