ATM regulation of IL-8 links oxidative stress to cancer cell migration and invasion
- PMID: 26030852
- PMCID: PMC4463759
- DOI: 10.7554/eLife.07270
ATM regulation of IL-8 links oxidative stress to cancer cell migration and invasion
Abstract
Ataxia-telangiectasia mutated (ATM) protein kinase regulates the DNA damage response (DDR) and is associated with cancer suppression. Here we report a cancer-promoting role for ATM. ATM depletion in metastatic cancer cells reduced cell migration and invasion. Transcription analyses identified a gene network, including the chemokine IL-8, regulated by ATM. IL-8 expression required ATM and was regulated by oxidative stress. IL-8 was validated as an ATM target by its ability to rescue cell migration and invasion defects in ATM-depleted cells. Finally, ATM-depletion in human breast cancer cells reduced lung tumors in a mouse xenograft model and clinical data validated IL-8 in lung metastasis. These findings provide insights into how ATM activation by oxidative stress regulates IL-8 to sustain cell migration and invasion in cancer cells to promote metastatic potential. Thus, in addition to well-established roles in tumor suppression, these findings identify a role for ATM in tumor progression.
Keywords: ATM; DNA damage; IL-8; ROS; cell biology; cell migration; chromosomes; genes; human; mouse.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
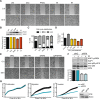
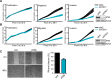
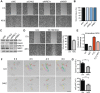




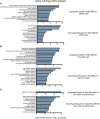
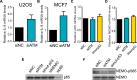
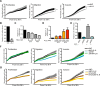

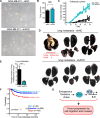
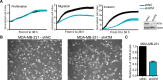

References
-
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, Parenti AR, Rosato A, Bicciato S, Balmain A, Piccolo S. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. - DOI - PubMed
-
- Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, Kusewitt D, Mills GB, Kastan MB, Walker CL. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proceedings of the National Academy of Sciences of USA. 2010;107:4153–4158. doi: 10.1073/pnas.0913860107. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

