Structural and kinetic characterization of recombinant 2-hydroxymuconate semialdehyde dehydrogenase from Pseudomonas putida G7
- PMID: 26032336
- PMCID: PMC4866608
- DOI: 10.1016/j.abb.2015.05.006
Structural and kinetic characterization of recombinant 2-hydroxymuconate semialdehyde dehydrogenase from Pseudomonas putida G7
Abstract
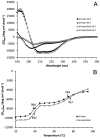
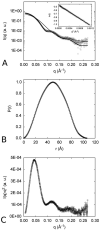
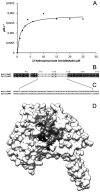
The first enzyme in the oxalocrotonate branch of the naphthalene-degradation lower pathway in Pseudomonas putida G7 is NahI, a 2-hydroxymuconate semialdehyde dehydrogenase which converts 2-hydroxymuconate semialdehyde to 2-hydroxymuconate in the presence of NAD(+). NahI is in family 8 (ALDH8) of the NAD(P)(+)-dependent aldehyde dehydrogenase superfamily. In this work, we report the cloning, expression, purification and preliminary structural and kinetic characterization of the recombinant NahI. The nahI gene was subcloned into a T7 expression vector and the enzyme was overexpressed in Escherichia coli ArcticExpress as a hexa-histidine-tagged fusion protein. After purification by affinity and size-exclusion chromatography, dynamic light scattering and small-angle X-ray scattering experiments were conducted to analyze the oligomeric state and the overall shape of the enzyme in solution. The protein is a tetramer in solution and has nearly perfect 222 point group symmetry. Protein stability and secondary structure content were evaluated by a circular dichroism spectroscopy assay under different thermal conditions. Furthermore, kinetic assays were conducted and, for the first time, KM (1.3±0.3μM) and kcat (0.9s(-1)) values were determined at presumed NAD(+) saturation. NahI is highly specific for its biological substrate and has no activity with salicylaldehyde, another intermediate in the naphthalene-degradation pathway.
Keywords: 2-Hydroxymuconate semialdehyde dehydrogenase; Kinetics; Naphthalene degradation; Pseudomonas putida G7; Structure.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

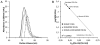

Similar articles
-
A combined approach for enhancing the stability of recombinant cis-dihydrodiol naphthalene dehydrogenase from Pseudomonas putida G7 allowed for the structural and kinetic characterization of the enzyme.Protein Expr Purif. 2017 Apr;132:50-59. doi: 10.1016/j.pep.2017.01.005. Epub 2017 Jan 9. Protein Expr Purif. 2017. PMID: 28089880
-
Expression, purification and preliminary crystallographic studies of NahF, a salicylaldehyde dehydrogenase from Pseudomonas putida G7 involved in naphthalene degradation.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Jan 1;68(Pt 1):93-7. doi: 10.1107/S174430911105038X. Epub 2011 Dec 24. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 22232182 Free PMC article.
-
Structure of formaldehyde dehydrogenase from Pseudomonas aeruginosa: the binary complex with the cofactor NAD+.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Sep;69(Pt 9):967-72. doi: 10.1107/S174430911302160X. Epub 2013 Aug 19. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23989142 Free PMC article.
-
[Structural and functional analysis of enzymes and their application to clinical analysis--study on Pseudomonas putida formaldehyde dehydrogenase].Yakugaku Zasshi. 2002 Oct;122(10):805-11. doi: 10.1248/yakushi.122.805. Yakugaku Zasshi. 2002. PMID: 12400161 Review. Japanese.
-
Conformational constraints in NAD analogs: implications for dehydrogenase binding and specificity.Adv Enzyme Regul. 2000;40:405-26. doi: 10.1016/s0065-2571(99)00056-4. Adv Enzyme Regul. 2000. PMID: 10828360 Review. No abstract available.
References
-
- Peng RH, et al. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol Rev. 2008;32(6):927–55. - PubMed
-
- Haritash AK, Kaushik CP. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater. 2009;169(1-3):1–15. - PubMed
-
- EPA Toxicological review of naphthalene. 2004.
-
- Singh S, et al. Bioremediation: environmental clean-up through pathway engineering. Current Opinion in Biotechnology. 2008;19:437–444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

