Atomic Structure of GRK5 Reveals Distinct Structural Features Novel for G Protein-coupled Receptor Kinases
- PMID: 26032409
- PMCID: PMC4543624
- DOI: 10.1074/jbc.M115.647297
Atomic Structure of GRK5 Reveals Distinct Structural Features Novel for G Protein-coupled Receptor Kinases
Abstract
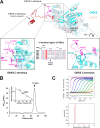
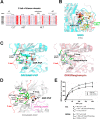
G protein-coupled receptor kinases (GRKs) are members of the protein kinase A, G, and C families (AGC) and play a central role in mediating G protein-coupled receptor phosphorylation and desensitization. One member of the family, GRK5, has been implicated in several human pathologies, including heart failure, hypertension, cancer, diabetes, and Alzheimer disease. To gain mechanistic insight into GRK5 function, we determined a crystal structure of full-length human GRK5 at 1.8 Å resolution. GRK5 in complex with the ATP analog 5'-adenylyl β,γ-imidodiphosphate or the nucleoside sangivamycin crystallized as a monomer. The C-terminal tail (C-tail) of AGC kinase domains is a highly conserved feature that is divided into three segments as follows: the C-lobe tether, the active-site tether (AST), and the N-lobe tether (NLT). This domain is fully resolved in GRK5 and reveals novel interactions with the nucleotide and N-lobe. Similar to other AGC kinases, the GRK5 AST is an integral part of the nucleotide-binding pocket, a feature not observed in other GRKs. The AST also mediates contact between the kinase N- and C-lobes facilitating closure of the kinase domain. The GRK5 NLT is largely displaced from its previously observed position in other GRKs. Moreover, although the autophosphorylation sites in the NLT are >20 Å away from the catalytic cleft, they are capable of rapid cis-autophosphorylation suggesting high mobility of this region. In summary, we provide a snapshot of GRK5 in a partially closed state, where structural elements of the kinase domain C-tail are aligned to form novel interactions to the nucleotide and N-lobe not previously observed in other GRKs.
Keywords: G protein-coupled receptor (GPCR); G protein-coupled receptor kinase; crystallography; phosphorylation; protein kinase; receptor regulation; signal transduction.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Krupnick J. G., Benovic J. L. (1998) The role of receptor kinases and arrestins in G protein-coupled receptor regulation. Annu. Rev. Pharmacol. Toxicol. 38, 289–319 - PubMed
-
- Premont R. T., Gainetdinov R. R. (2007) Physiological roles of G protein-coupled receptor kinases and arrestins. Annu. Rev. Physiol. 69, 511–534 - PubMed
-
- Martini J. S., Raake P., Vinge L. E., DeGeorge B. R., Jr., DeGeorge B., Jr., Chuprun J. K., Harris D. M., Gao E., Eckhart A. D., Pitcher J. A., Koch W. J. (2008) Uncovering G protein-coupled receptor kinase-5 as a histone deacetylase kinase in the nucleus of cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A. 105, 12457–12462 - PMC - PubMed
-
- Gold J. I., Martini J. S., Hullmann J., Gao E., Chuprun J. K., Lee L., Tilley D. G., Rabinowitz J. E., Bossuyt J., Bers D. M., Koch W. J. (2013) Nuclear translocation of cardiac G protein-coupled receptor kinase 5 downstream of select Gq-activating hypertrophic ligands is a calmodulin-dependent process. PLoS One 8, e57324. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

