Crystal Structure of G Protein-coupled Receptor Kinase 5 in Complex with a Rationally Designed Inhibitor
- PMID: 26032411
- PMCID: PMC4543626
- DOI: 10.1074/jbc.M115.647370
Crystal Structure of G Protein-coupled Receptor Kinase 5 in Complex with a Rationally Designed Inhibitor
Abstract
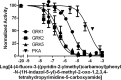
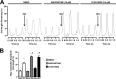
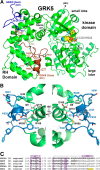
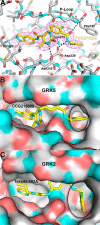
G protein-coupled receptor kinases (GRKs) regulate cell signaling by initiating the desensitization of active G protein-coupled receptors. The two most widely expressed GRKs (GRK2 and GRK5) play a role in cardiovascular disease and thus represent important targets for the development of novel therapeutic drugs. In the course of a GRK2 structure-based drug design campaign, one inhibitor (CCG215022) exhibited nanomolar IC50 values against both GRK2 and GRK5 and good selectivity against other closely related kinases such as GRK1 and PKA. Treatment of murine cardiomyocytes with CCG215022 resulted in significantly increased contractility at 20-fold lower concentrations than paroxetine, an inhibitor with more modest selectivity for GRK2. A 2.4 Å crystal structure of the GRK5·CCG215022 complex was determined and revealed that the inhibitor binds in the active site similarly to its parent compound GSK180736A. As designed, its 2-pyridylmethyl amide side chain occupies the hydrophobic subsite of the active site where it forms three additional hydrogen bonds, including one with the catalytic lysine. The overall conformation of the GRK5 kinase domain is similar to that of a previously determined structure of GRK6 in what is proposed to be its active state, but the C-terminal region of the enzyme adopts a distinct conformation. The kinetic properties of site-directed mutants in this region are consistent with the hypothesis that this novel C-terminal structure is representative of the membrane-bound conformation of the enzyme.
Keywords: G protein-coupled receptor kinase; crystallography; enzyme inhibitor; membrane targeting; plasma membrane; protein kinase; rational drug design; site-directed mutagenesis; x-ray crystallography.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Ferguson S. S. (2001) Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol. Rev. 53, 1–24 - PubMed
-
- Fredericks Z. L., Pitcher J. A., Lefkowitz R. J. (1996) Identification of the G protein-coupled receptor kinase phosphorylation sites in the human beta2-adrenergic receptor. J. Biol. Chem. 271, 13796–13803 - PubMed
-
- Liggett S. B., Ostrowski J., Chesnut L. C., Kurose H., Raymond J. R., Caron M. G., Lefkowitz R. J. (1992) Sites in the third intracellular loop of the α2A-adrenergic receptor confer short term agonist-promoted desensitization. Evidence for a receptor kinase-mediated mechanism. J. Biol. Chem. 267, 4740–4746 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- HL086865/HL/NHLBI NIH HHS/United States
- HL071818/HL/NHLBI NIH HHS/United States
- P01 HL108806/HL/NHLBI NIH HHS/United States
- R01 HL086865/HL/NHLBI NIH HHS/United States
- P60 DK020572/DK/NIDDK NIH HHS/United States
- P01 HL075443/HL/NHLBI NIH HHS/United States
- DK20572/DK/NIDDK NIH HHS/United States
- P01 HL108806 (PROJECT 3)/HL/NHLBI NIH HHS/United States
- R37 HL061690/HL/NHLBI NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- R01 HL071818/HL/NHLBI NIH HHS/United States
- P01 HL091799/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

