Complex Formation between Two Biosynthetic Enzymes Modifies the Allosteric Regulatory Properties of Both: AN EXAMPLE OF MOLECULAR SYMBIOSIS
- PMID: 26032422
- PMCID: PMC4505062
- DOI: 10.1074/jbc.M115.638700
Complex Formation between Two Biosynthetic Enzymes Modifies the Allosteric Regulatory Properties of Both: AN EXAMPLE OF MOLECULAR SYMBIOSIS
Abstract
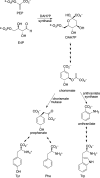
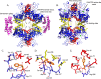
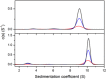
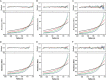
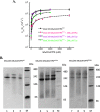
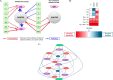
Allostery, where remote ligand binding alters protein function, is essential for the control of metabolism. Here, we have identified a highly sophisticated allosteric response that allows complex control of the pathway for aromatic amino acid biosynthesis in the pathogen Mycobacterium tuberculosis. This response is mediated by an enzyme complex formed by two pathway enzymes: chorismate mutase (CM) and 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (DAH7PS). Whereas both enzymes are active in isolation, the catalytic activity of both enzymes is enhanced, and in particular that of the much smaller CM is greatly enhanced (by 120-fold), by formation of a hetero-octameric complex between CM and DAH7PS. Moreover, on complex formation M. tuberculosis CM, which has no allosteric response on its own, acquires allosteric behavior to facilitate its own regulatory needs by directly appropriating and partly reconfiguring the allosteric machinery that provides a synergistic allosteric response in DAH7PS. Kinetic and analytical ultracentrifugation experiments demonstrate that allosteric binding of phenylalanine specifically promotes hetero-octameric complex dissociation, with concomitant reduction of CM activity. Together, DAH7PS and CM from M. tuberculosis provide exquisite control of aromatic amino acid biosynthesis, not only controlling flux into the start of the pathway, but also directing the pathway intermediate chorismate into either Phe/Tyr or Trp biosynthesis.
Keywords: 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase; Mycobacterium tuberculosis; TB; allosteric regulation; chorismate mutase; enzyme catalysis; oligomer; protein complex; protein-protein interaction; shikimate.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Perica T., Marsh J. A., Sousa F. L., Natan E., Colwell L. J., Ahnert S. E., Teichmann S. A. (2012) The emergence of protein complexes: quaternary structure, dynamics and allostery. Biochem. Soc. Trans. 40, 475–491 - PubMed
-
- McConkey G. A., Pinney J. W., Westhead D. R., Plueckhahn K., Fitzpatrick T. B., Macheroux P., Kappes B. (2004) Annotating the Plasmodium genome and the enigma of the shikimate pathway. Trends Parasitol. 20, 60–65 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials

