Quantitative analysis of commensal Escherichia coli populations reveals host-specific enterotypes at the intra-species level
- PMID: 26033772
- PMCID: PMC4554456
- DOI: 10.1002/mbo3.266
Quantitative analysis of commensal Escherichia coli populations reveals host-specific enterotypes at the intra-species level
Abstract
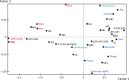
The primary habitat of the Escherichia coli species is the gut of warm-blooded vertebrates. The E. coli species is structured into four main phylogenetic groups A, B1, B2, and D. We estimated the relative proportions of these phylogroups in the feces of 137 wild and domesticated animals with various diets living in the Ile de France (Paris) region by real-time PCR. We distinguished three main clusters characterized by a particular abundance of two or more phylogroups within the E. coli animal commensal populations, which we called "enterocolitypes" by analogy with the enterotypes defined in the human gut microbiota at the genus level. These enterocolitypes were characterized by a dominant (>50%) B2, B1, or A phylogroup and were associated with different host species, diets, and habitats: wild and herbivorous species (wild rabbits and deer), domesticated herbivorous species (domesticated rabbits, horses, sheep, and cows), and omnivorous species (boar, pigs, and chickens), respectively. By analyzing retrospectively the data obtained using the same approach from 98 healthy humans living in Ile de France (Smati et al. 2013, Appl. Environ. Microbiol. 79, 5005-5012), we identified a specific human enterocolitype characterized by the dominant and/or exclusive (>90%) presence of phylogroup B2. We then compared B2 strains isolated from animals and humans, and revealed that human and animal strains differ regarding O-type and B2 subgroup. Moreover, two genes, sfa/foc and clbQ, were associated with the exclusive character of strains, observed only in humans. In conclusion, a complex network of interactions exists at several levels (genus and intra-species) within the intestinal microbiota.
Keywords: Commensal; Escherichia coli; enterotypes; microbiota; quantitative analysis.
© 2015 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures


References
-
- Baldy-Chudzik K, Mackiewicz P. Stosik M. Phylogenetic background, virulence gene profiles, and genomic diversity in commensal Escherichia coli isolated from ten mammal species living in one zoo. Vet. Microbiol. 2008;131:173–184. - PubMed
-
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

