A molecular description of cellulose biosynthesis
- PMID: 26034894
- PMCID: PMC4710354
- DOI: 10.1146/annurev-biochem-060614-033930
A molecular description of cellulose biosynthesis
Abstract
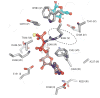
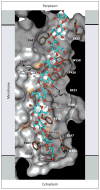
Cellulose is the most abundant biopolymer on Earth, and certain organisms from bacteria to plants and animals synthesize cellulose as an extracellular polymer for various biological functions. Humans have used cellulose for millennia as a material and an energy source, and the advent of a lignocellulosic fuel industry will elevate it to the primary carbon source for the burgeoning renewable energy sector. Despite the biological and societal importance of cellulose, the molecular mechanism by which it is synthesized is now only beginning to emerge. On the basis of recent advances in structural and molecular biology on bacterial cellulose synthases, we review emerging concepts of how the enzymes polymerize glucose molecules, how the nascent polymer is transported across the plasma membrane, and how bacterial cellulose biosynthesis is regulated during biofilm formation. Additionally, we review evolutionary commonalities and differences between cellulose synthases that modulate the nature of the cellulose product formed.
Keywords: biofilm; cellulose synthase; cyclic di-GMP; exopolysaccharide biosynthesis; membrane transport; processive glycosyltransferase.
Figures

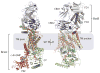

References
-
- Payen A. Mémoire sur la composition du tissu propre des plantes et du ligneux. C R Hebd Séances Acad Sci. 1838;7:1062–56.
-
- Klemm D, Heublein B, Fink HP, Bohn A. Cellulose: fascinating biopolymer and sustainable raw material. Angew Chem Int Ed. 2005;44:3358–93. - PubMed
-
- Staudinger H. Über Polymerisation. Ber Dtsch Chem Ges. 1920;53:1073–85.
-
- Wolfenden R, Lu X, Young G. Spontaneous hydrolysis of glycosides. J Am Chem Soc. 1998;120:6814–15.
-
- Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54:559–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

