Fecal Microbiota in Healthy Subjects Following Omnivore, Vegetarian and Vegan Diets: Culturable Populations and rRNA DGGE Profiling
- PMID: 26035837
- PMCID: PMC4452701
- DOI: 10.1371/journal.pone.0128669
Fecal Microbiota in Healthy Subjects Following Omnivore, Vegetarian and Vegan Diets: Culturable Populations and rRNA DGGE Profiling
Abstract
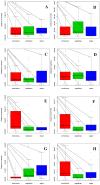
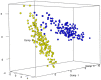
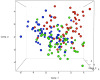
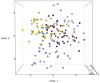
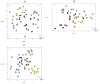
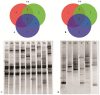
In this study, the fecal microbiota of 153 healthy volunteers, recruited from four different locations in Italy, has been studied by coupling viable counts, on different microbiological media, with ribosomal RNA Denaturing Gradient Gel Electrophoresis (rRNA-DGGE). The volunteers followed three different diets, namely omnivore, ovo-lacto-vegetarian and vegan. The results obtained from culture-dependent and -independent methods have underlined a high level of similarity of the viable fecal microbiota for the three investigated diets. The rRNA DGGE profiles were very complex and comprised a total number of bands that varied from 67 to 64 for the V3 and V9 regions of the 16S rRNA gene, respectively. Only a few bands were specific in/of all three diets, and the presence of common taxa associated with the dietary habits was found. As far as the viable counts are concerned, the high similarity of the fecal microbiota was once again confirmed, with only a few of the investigated groups showing significant differences. Interestingly, the samples grouped differently, according to the recruitment site, thus highlighting a higher impact of the food consumed by the volunteers in the specific geographical locations than that of the type of diet. Lastly, it should be mentioned that the fecal microbiota DGGE profiles obtained from the DNA were clearly separated from those produced using RNA, thus underlining a difference between the total and viable populations in the fecal samples.
Conflict of interest statement
Figures
References
-
- Foerster J, Maskarinec G, Reichardt N, Tett A, Narbad A, Blaut M, et al. The Influence of Whole Grain Products and Red Meat on Intestinal Microbiota Composition in Normal Weight Adults: A Randomized Crossover Intervention Trial. PLoS One. 2014;9:e109606 10.1371/journal.pone.0109606 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

