Dynamic Changes in ANGUSTIFOLIA3 Complex Composition Reveal a Growth Regulatory Mechanism in the Maize Leaf
- PMID: 26036253
- PMCID: PMC4498210
- DOI: 10.1105/tpc.15.00269
Dynamic Changes in ANGUSTIFOLIA3 Complex Composition Reveal a Growth Regulatory Mechanism in the Maize Leaf
Abstract
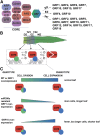
Most molecular processes during plant development occur with a particular spatio-temporal specificity. Thus far, it has remained technically challenging to capture dynamic protein-protein interactions within a growing organ, where the interplay between cell division and cell expansion is instrumental. Here, we combined high-resolution sampling of the growing maize (Zea mays) leaf with tandem affinity purification followed by mass spectrometry. Our results indicate that the growth-regulating SWI/SNF chromatin remodeling complex associated with ANGUSTIFOLIA3 (AN3) was conserved within growing organs and between dicots and monocots. Moreover, we were able to demonstrate the dynamics of the AN3-interacting proteins within the growing leaf, since copurified GROWTH-REGULATING FACTORs (GRFs) varied throughout the growing leaf. Indeed, GRF1, GRF6, GRF7, GRF12, GRF15, and GRF17 were significantly enriched in the division zone of the growing leaf, while GRF4 and GRF10 levels were comparable between division zone and expansion zone in the growing leaf. These dynamics were also reflected at the mRNA and protein levels, indicating tight developmental regulation of the AN3-associated chromatin remodeling complex. In addition, the phenotypes of maize plants overexpressing miRNA396a-resistant GRF1 support a model proposing that distinct associations of the chromatin remodeling complex with specific GRFs tightly regulate the transition between cell division and cell expansion. Together, our data demonstrate that advancing from static to dynamic protein-protein interaction analysis in a growing organ adds insights in how developmental switches are regulated.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures


References
-
- Antoni R., Gonzalez-Guzman M., Rodriguez L., Peirats-Llobet M., Pizzio G.A., Fernandez M.A., De Winne N., De Jaeger G., Dietrich D., Bennett M.J., Rodriguez P.L. (2013). PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 161: 931–941. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

