Identification of novel genes that regulate androgen receptor signaling and growth of androgen-deprived prostate cancer cells
- PMID: 26036626
- PMCID: PMC4537001
- DOI: 10.18632/oncotarget.3743
Identification of novel genes that regulate androgen receptor signaling and growth of androgen-deprived prostate cancer cells
Abstract
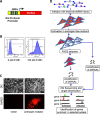
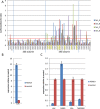
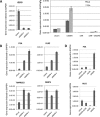
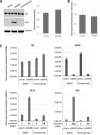
Prostate cancer progression to castration refractory disease is associated with anomalous transcriptional activity of the androgen receptor (AR) in an androgen-depleted milieu. To identify novel gene products whose downregulation transactivates AR in prostate cancer cells, we performed a screen of enzymatically-generated shRNA lenti-libraries selecting for transduced LNCaP cells with elevated expression of a fluorescent reporter gene under the control of an AR-responsive promoter. The shRNAs present in selected populations were analyzed using high-throughput sequencing to identify target genes. Highly enriched gene targets were then validated with siRNAs against selected genes, testing first for increased expression of luciferase from an AR-responsive promoter and then for altered expression of endogenous androgen-regulated genes in LNCaP cells. We identified 20 human genes whose silencing affected the expression of exogenous and endogenous androgen-responsive genes in prostate cancer cells grown in androgen-depleted medium. Knockdown of four of these genes upregulated the expression of endogenous AR targets and siRNAs targeting two of these genes (IGSF8 and RTN1) enabled androgen-independent proliferation of androgen-dependent cells. The effects of IGSF8 appear to be mediated through its interaction with a tetraspanin protein, CD9, previously implicated in prostate cancer progression. Remarkably, homozygous deletions of IGSF8 are found almost exclusively in prostate cancers but not in other cancer types. Our study shows that androgen independence can be achieved through the inhibition of specific genes and reveals a novel set of genes that regulate AR signaling in prostate cancers.
Keywords: IGSF8; androgen receptor; prostate cancer; shRNA; tumor progression.
Figures


References
-
- Zhang J, Thomas TZ, Kasper S, Matusik RJ. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology. 2000;141:4698–4710. - PubMed
-
- Kang Z, Pirskanen A, Janne OA, Palvimo JJ. Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. JBiolChem. 2002;277:48366–48371. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

