Bacteriophages limit the existence conditions for conjugative plasmids
- PMID: 26037122
- PMCID: PMC4453012
- DOI: 10.1128/mBio.00586-15
Bacteriophages limit the existence conditions for conjugative plasmids
Abstract
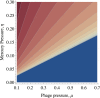
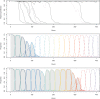
Bacteriophages are a major cause of bacterial mortality and impose strong selection on natural bacterial populations, yet their effects on the dynamics of conjugative plasmids have rarely been tested. We combined experimental evolution, mathematical modeling, and individual-based simulations to explain how the ecological and population genetics effects of bacteriophages upon bacteria interact to determine the dynamics of conjugative plasmids and their persistence. The ecological effects of bacteriophages on bacteria are predicted to limit the existence conditions for conjugative plasmids, preventing persistence under weak selection for plasmid accessory traits. Experiments showed that phages drove faster extinction of plasmids in environments where the plasmid conferred no benefit, but they also revealed more complex effects of phages on plasmid dynamics under these conditions, specifically, the temporary maintenance of plasmids at fixation followed by rapid loss. We hypothesized that the population genetic effects of bacteriophages, specifically, selection for phage resistance mutations, may have caused this. Further mathematical modeling and individual-based simulations supported our hypothesis, showing that conjugative plasmids may hitchhike with phage resistance mutations in the bacterial chromosome.
Importance: Conjugative plasmids are infectious loops of DNA capable of transmitting DNA between bacterial cells and between species. Because plasmids often carry extra genes that allow bacteria to live in otherwise-inhospitable environments, their dynamics are central to understanding bacterial adaptive evolution. The plasmid-bacterium interaction has typically been studied in isolation, but in natural bacterial communities, bacteriophages, viruses that infect bacteria, are ubiquitous. Using experiments, mathematical models, and computer simulations we show that bacteriophages drive plasmid dynamics through their ecological and evolutionary effects on bacteria and ultimately limit the conditions allowing plasmid existence. These results advance our understanding of bacterial adaptation and show that bacteriophages could be used to select against plasmids carrying undesirable traits, such as antibiotic resistance.
Copyright © 2015 Harrison et al.
Figures



References
-
- Macken CA, Levin SA, Waldstatter R. 1994. The dynamics of bacteria-plasmid systems. J Math Biol 32:123–145. doi:10.1007/BF00163028. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

