A sensitised RNAi screen reveals a ch-TOG genetic interaction network required for spindle assembly
- PMID: 26037491
- PMCID: PMC4453164
- DOI: 10.1038/srep10564
A sensitised RNAi screen reveals a ch-TOG genetic interaction network required for spindle assembly
Erratum in
-
Erratum: A sensitised RNAi screen reveals a ch-TOG genetic interaction network required for spindle assembly.Sci Rep. 2015 Jul 31;5:12384. doi: 10.1038/srep12384. Sci Rep. 2015. PMID: 26230530 Free PMC article. No abstract available.
Abstract
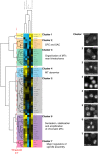
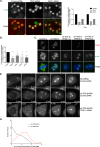
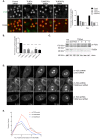
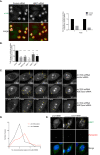
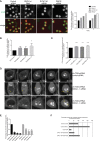
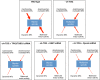
How multiple spindle assembly pathways are integrated to drive bipolar spindle assembly is poorly understood. We performed an image-based double RNAi screen to identify genes encoding Microtubule-Associated Proteins (MAPs) that interact with the highly conserved ch-TOG gene to regulate bipolar spindle assembly in human cells. We identified a ch-TOG centred network of genetic interactions which promotes centrosome-mediated microtubule polymerisation, leading to the incorporation of microtubules polymerised by all pathways into a bipolar structure [corrected]. Our genetic screen also reveals that ch-TOG maintains a dynamic microtubule population, in part, through modulating HSET activity. ch-TOG ensures that spindle assembly is robust to perturbation but sufficiently dynamic such that spindles can explore a diverse shape space in search of structures that can align chromosomes.
Figures

References
-
- Khodjakov A., Cole R.W., Oakley B.R. & Rieder C.L. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol 10, 59–67 (2000). - PubMed
-
- Wadsworth P. & Khodjakov A. E pluribus unum: towards a universal mechanism for spindle assembly. Trends Cell Biol 14, 413–419 (2004). - PubMed
-
- Luders J. & Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol 8, 161–167 (2007). - PubMed
-
- Luders J., Patel U.K. & Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol 8, 137–147 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

