Molecular evidence for interspecies transmission of H3N2pM/H3N2v influenza A viruses at an Ohio agricultural fair, July 2012
- PMID: 26038404
- PMCID: PMC3630945
- DOI: 10.1038/emi.2012.33
Molecular evidence for interspecies transmission of H3N2pM/H3N2v influenza A viruses at an Ohio agricultural fair, July 2012
Abstract
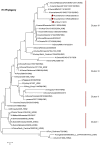
Evidence accumulating in 2011-2012 indicates that there is significant intra- and inter-species transmission of influenza A viruses at agricultural fairs, which has renewed interest in this unique human/swine interface. Six human cases of influenza A (H3N2) variant (H3N2v) virus infections were epidemiologically linked to swine exposure at fairs in the United States in 2011. In 2012, the number of H3N2v cases in the Midwest had exceeded 300 from early July to September, 2012. Prospective influenza A virus surveillance among pigs at Ohio fairs resulted in the detection of H3N2pM (H3N2 influenza A viruses containing the matrix (M) gene from the influenza A (H1N1) pdm09 virus). These H3N2pM viruses were temporally and spatially linked to several human H3N2v cases. Complete genomic analyses of these H3N2pM isolates demonstrated >99% nucleotide similarity to the H3N2v isolates recovered from human cases. Actions to mitigate the bidirectional interspecies transmission of influenza A virus between people and animals at agricultural fairs may be warranted.
Keywords: H3N2pM; fair; human H3N2v; influenza; swine.
Figures





References
-
- Easterday BC. The epidemiology and ecology of swine influenza as a zoonotic disease. Comp Immunol Microbiol Infect Dis. 1980;3:105–109. - PubMed
-
- Wentworth DE, McGregor MW, Macklin MD, Neumann V, Hinshaw VS. Transmission of swine influenza virus to humans after exposure to experimentally infected pigs. J Infect Dis. 1997;175:7–15. - PubMed
LinkOut - more resources
Full Text Sources
