Molecular biology and replication of hepatitis E virus
- PMID: 26038426
- PMCID: PMC3630916
- DOI: 10.1038/emi.2012.7
Molecular biology and replication of hepatitis E virus
Abstract
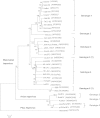
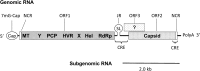
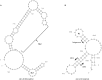
Hepatitis E virus (HEV), a single-stranded, positive-sense RNA virus, is responsible for acute hepatitis E epidemics in many developing countries, and the virus is also endemic in some industrialized countries. Hepatitis E is a recognized zoonotic disease, and several animal species, including pigs, are potential reservoirs for HEV. The genome of HEV contains three open reading frames (ORFs). ORF1 encodes the nonstructural proteins, ORF2 encodes the capsid protein, and ORF3 encodes a small multifunctional protein. The ORF2 and ORF3 proteins are translated from a single, bicistronic mRNA. The coding sequences for these two ORFs overlap each other, but neither overlaps with ORF1. Whereas the mechanisms underlying HEV replication are poorly understood, the construction of infectious viral clones, the identification of cell lines that support HEV replication, and the development of small animal models have allowed for more detailed study of the virus. As result of these advances, recently, our understanding of viral entry, genomic replication and viral egress has improved. Furthermore, the determination of the T=1 and T=3 structure of HEV virus-like particles has furthered our understanding of the replication of HEV. This article reviews the latest developments in the molecular biology of HEV with an emphasis on the genomic organization, the expression and function of genes, and the structure and replication of HEV.
Keywords: animal model; hepatitis E; hepatitis E virus; molecular biology; replication; zoonosis.
Figures

References
-
- Emerson SU, Purcell RH.Hepatitis E VirusIn: Knipe DM, Howley PM, editors. Fields Virology. 5th ed Philadelphia, PA; Lippincott-Raven Publishers; 20073048–3059.
-
- Meng XJ.Hepatitis E Virus (Hepevirus)In: Mahy BWJ, Regenmortel MHV, editors. Encyclopedia of Virology. 3rd ed Oxford; Academic Press; 2008377–383.
-
- Dalton HR, Bendall R, Ijaz S, Banks M. Hepatitis E: an emerging infection in developed countries. Lancet Infect Dis. 2008;8:698–709. - PubMed
-
- FitzSimons D, Hendrickx G, Vorsters A, van Damme P. Hepatitis A and E: update on prevention and epidemiology. Vaccine. 2010;28:583–588. - PubMed
-
- Mushahwar IK. Hepatitis E virus: molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. J Med Virol. 2008;80:646–658. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
