Hepatitis B virus reverse transcriptase: diverse functions as classical and emerging targets for antiviral intervention
- PMID: 26038488
- PMCID: PMC3820986
- DOI: 10.1038/emi.2013.56
Hepatitis B virus reverse transcriptase: diverse functions as classical and emerging targets for antiviral intervention
Abstract
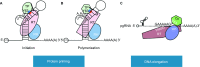
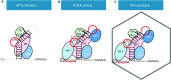
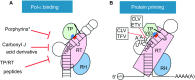
Hepatitis B virus (HBV) infection remains a global health problem with over 350 million chronically infected, causing an increased risk of cirrhosis and hepatocellular carcinoma. Current antiviral chemotherapy for HBV infection include five nucleos(t)ide analog reverse transcriptase inhibitors (NRTIs) that all target one enzymatic activity, DNA strand elongation, of the HBV polymerase (HP), a specialized reverse transcriptase (RT). NRTIs are not curative and long-term treatment is associated with toxicity and the emergence of drug resistant viral mutations, which can also result in vaccine escape. Recent studies on the multiple functions of HP have provided important mechanistic insights into its diverse roles during different stages of viral replication, including interactions with viral pregenomic RNA, RNA packaging into nucleocapsids, protein priming, minus- and plus-strand viral DNA synthesis, RNase H-mediated degradation of viral RNA, as well as critical host interactions that regulate the multiple HP functions. These diverse functions provide ample opportunities to develop novel HP-targeted antiviral treatments that should contribute to curing chronic HBV infection.
Keywords: Hepadnavirus; RNA packaging; RNase H; hepatitis B virus; protein priming; reverse transcriptase; review.
Figures

References
-
- Seeger C, Mason WS, Zoulim F.HepadnavirusesIn: KnipeDM, HowleyPM (eds.)Fields virology Philadelphia, PA; Lippincott Williams & Wilkins; 20072977–3029.
-
- Hu J, Seeger C. Expression and characterization of hepadnavirus reverse transcriptases. Methods Enzymol. 1996;275:195–208. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
