Exploring the dynamics of cell processes through simulations of fluorescence microscopy experiments
- PMID: 26039162
- PMCID: PMC4457496
- DOI: 10.1016/j.bpj.2015.04.014
Exploring the dynamics of cell processes through simulations of fluorescence microscopy experiments
Abstract
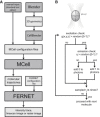
Fluorescence correlation spectroscopy (FCS) methods are powerful tools for unveiling the dynamical organization of cells. For simple cases, such as molecules passively moving in a homogeneous media, FCS analysis yields analytical functions that can be fitted to the experimental data to recover the phenomenological rate parameters. Unfortunately, many dynamical processes in cells do not follow these simple models, and in many instances it is not possible to obtain an analytical function through a theoretical analysis of a more complex model. In such cases, experimental analysis can be combined with Monte Carlo simulations to aid in interpretation of the data. In response to this need, we developed a method called FERNET (Fluorescence Emission Recipes and Numerical routines Toolkit) based on Monte Carlo simulations and the MCell-Blender platform, which was designed to treat the reaction-diffusion problem under realistic scenarios. This method enables us to set complex geometries of the simulation space, distribute molecules among different compartments, and define interspecies reactions with selected kinetic constants, diffusion coefficients, and species brightness. We apply this method to simulate single- and multiple-point FCS, photon-counting histogram analysis, raster image correlation spectroscopy, and two-color fluorescence cross-correlation spectroscopy. We believe that this new program could be very useful for predicting and understanding the output of fluorescence microscopy experiments.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Dertinger T., Pacheco V., Enderlein J. Two-focus fluorescence correlation spectroscopy: a new tool for accurate and absolute diffusion measurements. ChemPhysChem. 2007;8:433–443. - PubMed
-
- Burkhardt M., Schwille P. Electron multiplying CCD based detection for spatially resolved fluorescence correlation spectroscopy. Opt. Express. 2006;14:5013–5020. - PubMed
-
- Bacia K., Kim S.A., Schwille P. Fluorescence cross-correlation spectroscopy in living cells. Nat. Methods. 2006;3:83–89. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

