A cross comparison of technologies for the detection of microRNAs in clinical FFPE samples of hepatoblastoma patients
- PMID: 26039282
- PMCID: PMC4453922
- DOI: 10.1038/srep10438
A cross comparison of technologies for the detection of microRNAs in clinical FFPE samples of hepatoblastoma patients
Erratum in
-
Erratum: A cross comparison of technologies for the detection of microRNAs in clinical FFPE samples of hepatoblastoma patients.Sci Rep. 2015 Sep 2;5:13505. doi: 10.1038/srep13505. Sci Rep. 2015. PMID: 26328756 Free PMC article. No abstract available.
Abstract
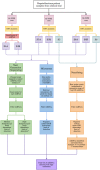
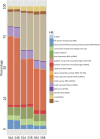
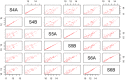
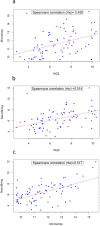
Although formalin fixed paraffin embedded (FFPE) tissue is a major biological source in cancer research, it is challenging to work with due to macromolecular fragmentation and nucleic acid crosslinking. Therefore, it is important to characterise the quality of data that can be obtained from FFPE samples. We have compared three independent platforms (next generation sequencing, microarray and NanoString) for profiling microRNAs (miRNAs) using clinical FFPE samples from hepatoblastoma (HB) patients. The number of detected miRNAs ranged from 228 to 345 (median = 294) using the next generation sequencing platform, whereas 79 to 125 (median = 112) miRNAs were identified using microarrays in three HB samples, including technical replicates. NanoString identified 299 to 372 miRNAs in two samples. Between the platforms, we observed high reproducibility and significant levels of shared detection. However, for commonly detected miRNAs, a strong correlation between platforms was not observed. Analysis of 10 additional HB samples with NanoString identified significantly overlapping miRNA expression profiles, and an alternative pattern was identified in a poorly differentiated HB with an aggressive phenotype. This investigation serves as a roadmap for future studies investigating miRNA expression in clinical FFPE samples, and as a guideline for the selection of an appropriate platform.
Figures



References
-
- Krek A. et al. Combinatorial microRNA target predictions. Nat Genet 37, 495–500 (2005). - PubMed
-
- Lewis B. P., Burge C. B. & Bartel D. P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell 120, 15–20 (2005). - PubMed
-
- Esquela-Kerscher A. & Slack F. J. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6, 259–269 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

