Non-coding RNAs: Epigenetic regulators of bone development and homeostasis
- PMID: 26039869
- PMCID: PMC6095476
- DOI: 10.1016/j.bone.2015.05.026
Non-coding RNAs: Epigenetic regulators of bone development and homeostasis
Abstract
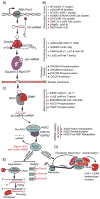
Non-coding RNAs (ncRNAs) have evolved in eukaryotes as epigenetic regulators of gene expression. The most abundant regulatory ncRNAs are the 20-24 nt small microRNAs (miRNAs) and long non-coding RNAs (lncRNAs, <200 nt). Each class of ncRNAs operates through distinct mechanisms, but their pathways to regulating gene expression are interrelated in ways that are just being recognized. While the importance of lncRNAs in epigenetic control of transcription, developmental processes and human traits is emerging, the identity of lncRNAs in skeletal biology is scarcely known. However, since the first profiling studies of miRNA at stages during osteoblast and osteoclast differentiation, over 1100 publications related to bone biology and pathologies can be found, as well as many recent comprehensive reviews summarizing miRNA in skeletal cells. Delineating the activities and targets of specific miRNAs regulating differentiation of osteogenic and resorptive bone cells, coupled with in vivo gain- and loss-of-function studies, discovered unique mechanisms that support bone development and bone homeostasis in adults. We present here "guiding principles" for addressing biological control of bone tissue formation by ncRNAs. This review emphasizes recent advances in understanding regulation of the process of miRNA biogenesis that impact on osteogenic lineage commitment, transcription factors and signaling pathways. Also discussed are the approaches to be pursued for an understanding of the role of lncRNAs in bone and the challenges in addressing their multiple and complex functions. Based on new knowledge of epigenetic control of gene expression to be gained for ncRNA regulation of the skeleton, new directions for translating the miRNAs and lncRNAs into therapeutic targets for skeletal disorders are possible. This article is part of a Special Issue entitled Epigenetics and Bone.
Keywords: LncRNAs; MicroRNA; Osteoblasts; miRNA biogenesis.
Published by Elsevier Inc.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

