Improved detection of Escherichia coli and coliform bacteria by multiplex PCR
- PMID: 26040540
- PMCID: PMC4453288
- DOI: 10.1186/s12896-015-0168-2
Improved detection of Escherichia coli and coliform bacteria by multiplex PCR
Abstract
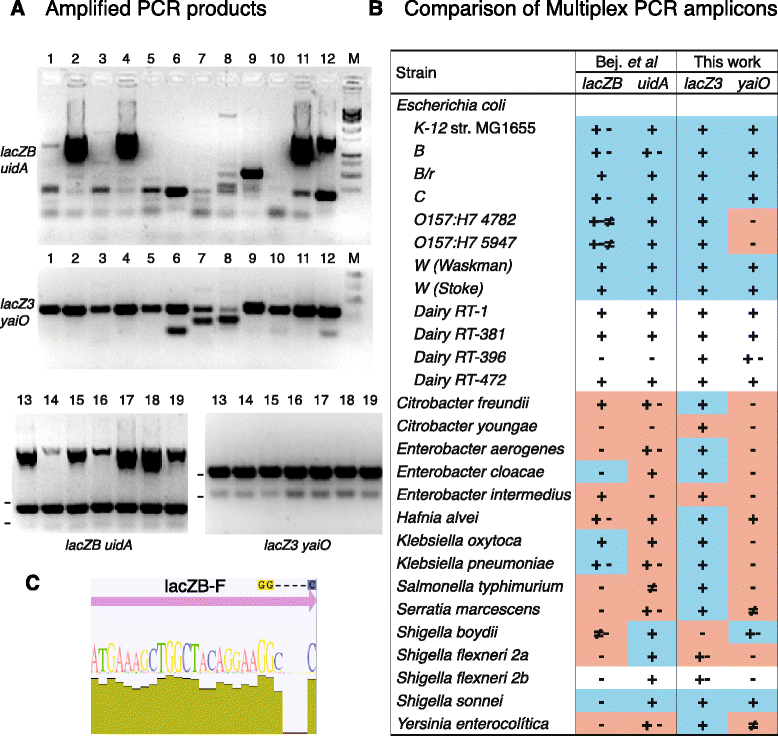
Background: The presence of coliform bacteria is routinely assessed to establish the microbiological safety of water supplies and raw or processed foods. Coliforms are a group of lactose-fermenting Enterobacteriaceae, which most likely acquired the lacZ gene by horizontal transfer and therefore constitute a polyphyletic group. Among this group of bacteria is Escherichia coli, the pathogen that is most frequently associated with foodborne disease outbreaks and is often identified by β-glucuronidase enzymatic activity or by the redundant detection of uidA by PCR. Because a significant fraction of essential E. coli genes are preserved throughout the bacterial kingdom, alternative oligonucleotide primers for specific E. coli detection are not easily identified.
Results: In this manuscript, two strategies were used to design oligonucleotide primers with differing levels of specificity for the simultaneous detection of total coliforms and E. coli by multiplex PCR. A consensus sequence of lacZ and the orphan gene yaiO were chosen as targets for amplification, yielding 234 bp and 115 bp PCR products, respectively.
Conclusions: The assay designed in this work demonstrated superior detection ability when tested with lab collection and dairy isolated lactose-fermenting strains. While lacZ amplicons were found in a wide range of coliforms, yaiO amplification was highly specific for E. coli. Additionally, yaiO detection is non-redundant with enzymatic methods.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

