ProCARs: Progressive Reconstruction of Ancestral Gene Orders
- PMID: 26040958
- PMCID: PMC4460626
- DOI: 10.1186/1471-2164-16-S5-S6
ProCARs: Progressive Reconstruction of Ancestral Gene Orders
Abstract
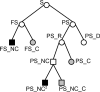
Background: In the context of ancestral gene order reconstruction from extant genomes, there exist two main computational approaches: rearrangement-based, and homology-based methods. The rearrangement-based methods consist in minimizing a total rearrangement distance on the branches of a species tree. The homology-based methods consist in the detection of a set of potential ancestral contiguity features, followed by the assembling of these features into Contiguous Ancestral Regions (CARs).
Results: In this paper, we present a new homology-based method that uses a progressive approach for both the detection and the assembling of ancestral contiguity features into CARs. The method is based on detecting a set of potential ancestral adjacencies iteratively using the current set of CARs at each step, and constructing CARs progressively using a 2-phase assembling method.
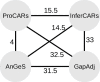
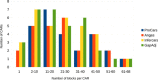
Conclusion: We show the usefulness of the method through a reconstruction of the boreoeutherian ancestral gene order, and a comparison with three other homology-based methods: AnGeS, InferCARs and GapAdj. The program, written in Python, and the dataset used in this paper are available at http://bioinfo.lifl.fr/procars/.
Figures




References
-
- Bergeron A, Blanchette M, Chateau A, Chauve C. Reconstructing ancestral gene orders using conserved intervals. Lecture Notes in Computer Science. 2004;3240:14–25. doi: 10.1007/978-3-540-30219-3_2. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

