The Chromatin Remodeling Protein Bptf Promotes Posterior Neuroectodermal Fate by Enhancing Smad2-Activated wnt8a Expression
- PMID: 26041917
- PMCID: PMC6605332
- DOI: 10.1523/JNEUROSCI.0377-15.2015
The Chromatin Remodeling Protein Bptf Promotes Posterior Neuroectodermal Fate by Enhancing Smad2-Activated wnt8a Expression
Abstract
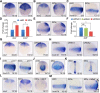
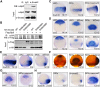
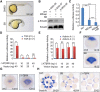
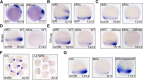
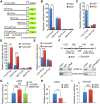
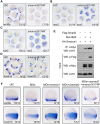
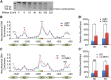
During vertebrate embryogenesis, the neuroectoderm is induced from dorsal ectoderm and then partitioned into anterior and posterior neuroectodermal domains by posteriorizing signals, such as Wnt and fibroblast growth factor. However, little is known about epigenetic regulation of posteriorizing gene expression. Here, we report a requirement of the chromatin remodeling protein Bptf for neuroectodermal posteriorization in zebrafish embryos. Knockdown of bptf leads to an expansion of the anterior neuroectoderm at the expense of the posterior ectoderm. Bptf functionally and physically interacts with p-Smad2, which is activated by non-Nodal TGF-β signaling, to promote the expression of wnt8a, a critical gene for neural posteriorization. Bptf and Smad2 directly bind to and activate the wnt8a promoter through recruiting NURF remodeling complex. When bptf function or TGF-β signal transduction is inhibited, the nucleosome density on the wnt8a promoter is increased. We propose that Bptf and TGF-β/Smad2 mediate nucleosome remodeling to regulate wnt8a expression and hence neural posteriorization.
Keywords: bptf; neural posteriorization; nucleosome remodeling; smad2; wnt8a.
Copyright © 2015 the authors 0270-6474/15/358493-14$15.00/0.
Figures


References
-
- Brand T, MacLellan WR, Schneider MD. A dominant-negative receptor for type beta transforming growth factors created by deletion of the kinase domain. J Biol Chem. 1993;268:11500–11503. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
