Transcriptome Characterization of Cymbidium sinense 'Dharma' Using 454 Pyrosequencing and Its Application in the Identification of Genes Associated with Leaf Color Variation
- PMID: 26042676
- PMCID: PMC4456352
- DOI: 10.1371/journal.pone.0128592
Transcriptome Characterization of Cymbidium sinense 'Dharma' Using 454 Pyrosequencing and Its Application in the Identification of Genes Associated with Leaf Color Variation
Abstract
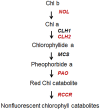
The highly variable leaf color of Cymbidium sinense significantly improves its horticultural and economic value, and makes it highly desirable in the flower markets in China and Southeast Asia. However, little is understood about the molecular mechanism underlying leaf-color variations. In this study, we found the content of photosynthetic pigments, especially chlorophyll degradation metabolite in the leaf-color mutants is distinguished significantly from that in the wild type of Cymbidium sinense 'Dharma'. To further determine the candidate genes controlling leaf-color variations, we first sequenced the global transcriptome using 454 pyrosequencing. More than 0.7 million expressed sequence tags (ESTs) with an average read length of 445.9 bp were generated and assembled into 103,295 isotigs representing 68,460 genes. Of these isotigs, 43,433 were significantly aligned to known proteins in the public database, of which 29,299 could be categorized into 42 functional groups in the gene ontology system, 10,079 classified into 23 functional classifications in the clusters of orthologous groups system, and 23,092 assigned to 139 clusters of specific metabolic pathways in the Kyoto Encyclopedia of Genes and Genomes. Among these annotations, 95 isotigs were designated as involved in chlorophyll metabolism. On this basis, we identified 16 key enzyme-encoding genes in the chlorophyll metabolism pathway, the full length cDNAs and expressions of which were further confirmed. Expression pattern indicated that the key enzyme-encoding genes for chlorophyll degradation were more highly expressed in the leaf color mutants, as was consistent with their lower chlorophyll contents. This study is the first to supply an informative 454 EST dataset for Cymbidium sinense 'Dharma' and to identify original leaf color-associated genes, which provide important resources to facilitate gene discovery for molecular breeding, marketable trait discovery, and investigating various biological process in this species.
Conflict of interest statement
Figures







References
-
- Liu ZJ, Chen SC, Ru ZZ, Chen LJ, editors (2006) Chinese Cymbidium plants. Beijing: Science Press.
-
- Huang J, Dai S (1998) The numerical taxonomy of Chinese Cymbidium . Journal of Beijing Forestry University 20: 38–43.
-
- Xu Y, Teo LL, Zhou J, Kumar PP, Yu H (2006) Floral organ identity genes in the orchid Dendrobium crumenatum . Plant J 46: 54–68. - PubMed
-
- Yukawa T, Stern WL (2002) Comparative vegetative anatomy and systematics of Cymbidium (Cymbidieae: Orchidaceae). Bot J Linn Soc 138: 383–419.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

