CRISPR-Cas9-mediated gene knockout in primary human airway epithelial cells reveals a proinflammatory role for MUC18
- PMID: 26043872
- PMCID: PMC4600011
- DOI: 10.1038/gt.2015.53
CRISPR-Cas9-mediated gene knockout in primary human airway epithelial cells reveals a proinflammatory role for MUC18
Abstract
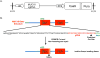
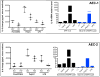
Targeted knockout of genes in primary human cells using CRISPR-Cas9-mediated genome-editing represents a powerful approach to study gene function and to discern molecular mechanisms underlying complex human diseases. We used lentiviral delivery of CRISPR-Cas9 machinery and conditional reprogramming culture methods to knockout the MUC18 gene in human primary nasal airway epithelial cells (AECs). Massively parallel sequencing technology was used to confirm that the genome of essentially all cells in the edited AEC populations contained coding region insertions and deletions (indels). Correspondingly, we found mRNA expression of MUC18 was greatly reduced and protein expression was absent. Characterization of MUC18 knockout cell populations stimulated with TLR2, 3 and 4 agonists revealed that IL-8 (a proinflammatory chemokine) responses of AECs were greatly reduced in the absence of functional MUC18 protein. Our results show the feasibility of CRISPR-Cas9-mediated gene knockouts in AEC culture (both submerged and polarized), and suggest a proinflammatory role for MUC18 in airway epithelial response to bacterial and viral stimuli.
Conflict of interest statement
Figures




References
-
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428(6981):431–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

