Structural Basis of Clade-specific Engagement of SAMHD1 (Sterile α Motif and Histidine/Aspartate-containing Protein 1) Restriction Factors by Lentiviral Viral Protein X (Vpx) Virulence Factors
- PMID: 26045556
- PMCID: PMC4505041
- DOI: 10.1074/jbc.M115.665513
Structural Basis of Clade-specific Engagement of SAMHD1 (Sterile α Motif and Histidine/Aspartate-containing Protein 1) Restriction Factors by Lentiviral Viral Protein X (Vpx) Virulence Factors
Abstract
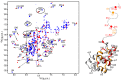
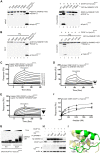
Sterile α motif (SAM) and histidine/aspartate (HD)-containing protein 1 (SAMHD1) restricts human/simian immunodeficiency virus infection in certain cell types and is counteracted by the virulence factor Vpx. Current evidence indicates that Vpx recruits SAMHD1 to the Cullin4-Ring Finger E3 ubiquitin ligase (CRL4) by facilitating an interaction between SAMHD1 and the substrate receptor DDB1- and Cullin4-associated factor 1 (DCAF1), thereby targeting SAMHD1 for proteasome-dependent down-regulation. Host-pathogen co-evolution and positive selection at the interfaces of host-pathogen complexes are associated with sequence divergence and varying functional consequences. Two alternative interaction interfaces are used by SAMHD1 and Vpx: the SAMHD1 N-terminal tail and the adjacent SAM domain or the C-terminal tail proceeding the HD domain are targeted by different Vpx variants in a unique fashion. In contrast, the C-terminal WD40 domain of DCAF1 interfaces similarly with the two above complexes. Comprehensive biochemical and structural biology approaches permitted us to delineate details of clade-specific recognition of SAMHD1 by lentiviral Vpx proteins. We show that not only the SAM domain but also the N-terminal tail engages in the DCAF1-Vpx interaction. Furthermore, we show that changing the single Ser-52 in human SAMHD1 to Phe, the residue found in SAMHD1 of Red-capped monkey and Mandrill, allows it to be recognized by Vpx proteins of simian viruses infecting those primate species, which normally does not target wild type human SAMHD1 for degradation.
Keywords: SAM domain and HD domain-containing protein 1 (SAMHD1); SIV; X-ray crystallography; human immunodeficiency virus (HIV); nuclear magnetic resonance (NMR); restriction factor; ubiquitylation (ubiquitination).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. (2004) The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

