The transcription factor Swi4 is target for PKA regulation of cell size at the G1 to S transition in Saccharomyces cerevisiae
- PMID: 26046481
- PMCID: PMC4614678
- DOI: 10.1080/15384101.2015.1055997
The transcription factor Swi4 is target for PKA regulation of cell size at the G1 to S transition in Saccharomyces cerevisiae
Abstract
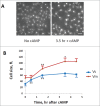
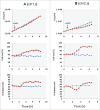
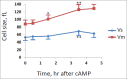
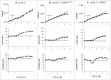
To investigate the specific target of PKA in the regulation of cell cycle progression and cell size we developed a new approach using the yeast strain GG104 bearing a deletion in adenylate cyclase gene and permeable to cAMP ( cyr1Δ, pde2Δ, msn2Δ, msn4Δ). In this strain the PKA activity is absent and can be activated by addition of cAMP in the medium, without any other change of the growth conditions. In the present work we show that the activation of PKA by exogenous cAMP in the GG104 strain exponentially growing in glucose medium caused a marked increase of cell size and perturbation of cell cycle with a transient arrest of cells in G1, followed by an accumulation of cells in G2/M phase with a minimal change in the growth rate. Deletion of CLN1 gene, but not of CLN2, abolished the transient G1 phase arrest. Consistently we found that PKA activation caused a transcriptional repression of CLN1 gene. Transcription of CLN1 is controlled by SBF and MBF dual-regulated promoter. We found that also the deletion of SWI4 gene abolished the transient G1 arrest suggesting that Swi4 is a target responsible for PKA modulation of G1/S phase transition. We generated a SWI4 allele mutated in the consensus site for PKA (Swi4(S159A)) and we found that expression of Swi4(S159A) protein in the GG104-Swi4Δ strain did not restore the transient G1 arrest induced by PKA activation, suggesting that Swi4 phosphorylation by PKA regulates CLN1 gene expression and G1/S phase transition.
Keywords: Cln1; Cyclic AMP; G1 cyclins; Mitosis; SBF; budding yeast.
Figures



References
-
- Pringle JR, Hartwell LH. The Saccharomyces cerevisiae cell cycle In: Strathern JN, Jones EW, Broach JR, editors The molecular biology of the yeast Saccharomyces cerevisiae: life cycle and inheritance. Cold Spring Harbor: New York, NY, USA: Cold Spring Harbor Laboratory; 1981; 97-142.
-
- Wagner MV, Smolka MB, de Bruin RAM, Zhou H, Wittenberg C, Dowdy SF. Whi5 regulation by site specific CDK-phosphorylation in Saccharomyces cerevisiae. PLoS One 2009; 4:e4300; PMID:19172996; http://dx.doi.org/10.1371/journal.pone.0004300 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
