Krt19(+)/Lgr5(-) Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine
- PMID: 26046762
- PMCID: PMC4457942
- DOI: 10.1016/j.stem.2015.04.013
Krt19(+)/Lgr5(-) Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine
Abstract
Epithelium of the colon and intestine are renewed every 3 days. In the intestine there are at least two principal stem cell pools. The first contains rapid cycling crypt-based columnar (CBC) Lgr5(+) cells, and the second is composed of slower cycling Bmi1-expressing cells at the +4 position above the crypt base. In the colon, however, the identification of Lgr5(-) stem cell pools has proven more challenging. Here, we demonstrate that the intermediate filament keratin-19 (Krt19) marks long-lived, radiation-resistant cells above the crypt base that generate Lgr5(+) CBCs in the colon and intestine. In colorectal cancer models, Krt19(+) cancer-initiating cells are also radioresistant, while Lgr5(+) stem cells are radiosensitive. Moreover, Lgr5(+) stem cells are dispensable in both the normal and neoplastic colonic epithelium, as ablation of Lgr5(+) stem cells results in their regeneration from Krt19-expressing cells. Thus, Krt19(+) stem cells are a discrete target relevant for cancer therapy.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors do not have any conflicts of interests to declare.
Figures

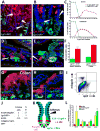
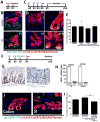
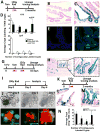
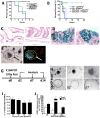
References
-
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. - PubMed
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- Brembeck FH, Moffett J, Wang TC, Rustgi AK. The keratin 19 promoter is potent for cell-specific targeting of genes in transgenic mice. Gastroenterology. 2001;120:1720–1728. - PubMed
-
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. - PubMed
-
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

