Effective Optimization of Antibody Affinity by Phage Display Integrated with High-Throughput DNA Synthesis and Sequencing Technologies
- PMID: 26046845
- PMCID: PMC4457833
- DOI: 10.1371/journal.pone.0129125
Effective Optimization of Antibody Affinity by Phage Display Integrated with High-Throughput DNA Synthesis and Sequencing Technologies
Abstract
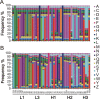
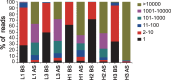
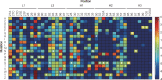
Phage display technology has been widely used for antibody affinity maturation for decades. The limited library sequence diversity together with excessive redundancy and labour-consuming procedure for candidate identification are two major obstacles to widespread adoption of this technology. We hereby describe a novel library generation and screening approach to address the problems. The approach started with the targeted diversification of multiple complementarity determining regions (CDRs) of a humanized anti-ErbB2 antibody, HuA21, with a small perturbation mutagenesis strategy. A combination of three degenerate codons, NWG, NWC, and NSG, were chosen for amino acid saturation mutagenesis without introducing cysteine and stop residues. In total, 7,749 degenerate oligonucleotides were synthesized on two microchips and released to construct five single-chain antibody fragment (scFv) gene libraries with 4 x 10(6) DNA sequences. Deep sequencing of the unselected and selected phage libraries using the Illumina platform allowed for an in-depth evaluation of the enrichment landscapes in CDR sequences and amino acid substitutions. Potent candidates were identified according to their high frequencies using NGS analysis, by-passing the need for the primary screening of target-binding clones. Furthermore, a subsequent library by recombination of the 10 most abundant variants from four CDRs was constructed and screened, and a mutant with 158-fold increased affinity (Kd = 25.5 pM) was obtained. These results suggest the potential application of the developed methodology for optimizing the binding properties of other antibodies and biomolecules.
Conflict of interest statement
Figures



References
-
- Wark KL, Hudson PJ. Latest technologies for the enhancement of antibody affinity. Adv Drug Deliv Rev. 2006; 58: 657–670. - PubMed
-
- Martineau P. Error-prone polymerase chain reaction for modification of scFvs. Methods Mol Biol. 2002; 178: 287–294. - PubMed
-
- Low NM, Holliger P, Winter G. Mimicking somatic hypermutation: Affinity maturation of antibodies displayed on bacteriophage using a bacterial mutator strain. J Mol Biol. 1996; 260: 359–368. - PubMed
-
- Irving RA, Kortt AA, Hudson PJ. Affinity maturation of recombinant antibodies using E-coli mutator cells. Immunotechnology.1996; 2: 127–143. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

