Refining the nuclear auxin response pathway through structural biology
- PMID: 26048079
- PMCID: PMC4618177
- DOI: 10.1016/j.pbi.2015.05.007
Refining the nuclear auxin response pathway through structural biology
Abstract
Auxin is a key regulator of plant growth and development. Classical molecular and genetic techniques employed over the past 20 years identified the major players in auxin-mediated gene expression and suggest a canonical auxin response pathway. In recent years, structural and biophysical studies clarified the molecular details of auxin perception, the recognition of DNA by auxin transcription factors, and the interaction of auxin transcription factors with repressor proteins. These studies refine the auxin signal transduction model and raise new questions that increase the complexity of auxin signaling.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
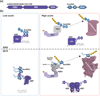
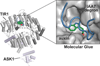
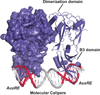

References
-
- Peer WA. From perception to attenuation: auxin signalling and responses. Curr Opin Plant Biol. 2013;16:561–568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

