Dynamics of the discovery process of protein-protein interactions from low content studies
- PMID: 26048415
- PMCID: PMC4456804
- DOI: 10.1186/s12918-015-0173-z
Dynamics of the discovery process of protein-protein interactions from low content studies
Abstract
Background: Thousands of biological and biomedical investigators study of the functional role of single genes and their protein products in normal physiology and in disease. The findings from these studies are reported in research articles that stimulate new research. It is now established that a complex regulatory networks's is controlling human cellular fate, and this community of researchers are continually unraveling this network topology. Attempts to integrate results from such accumulated knowledge resulted in literature-based protein-protein interaction networks (PPINs) and pathway databases. These databases are widely used by the community to analyze new data collected from emerging genome-wide studies with the assumption that the data within these literature-based databases is the ground truth and contain no biases. While suspicion for research focus biases is growing, a concrete proof for it is still missing. It is difficult to prove because the real PPINs are mostly unknown.
Results: Here we analyzed the longitudinal discovery process of literature-based mammalian and yeast PPINs to observe that these networks are discovered non-uniformly. The pattern of discovery is related to a theoretical concept proposed by Kauffman called "expanding the adjacent possible". We introduce a network discovery model which explicitly includes the space of possibilities in the form of a true underlying PPIN.
Conclusions: Our model strongly suggests that research focus biases exist in the observed discovery dynamics of these networks. In summary, more care should be placed when using PPIN databases for analysis of newly acquired data, and when considering prior knowledge when designing new experiments.
Figures


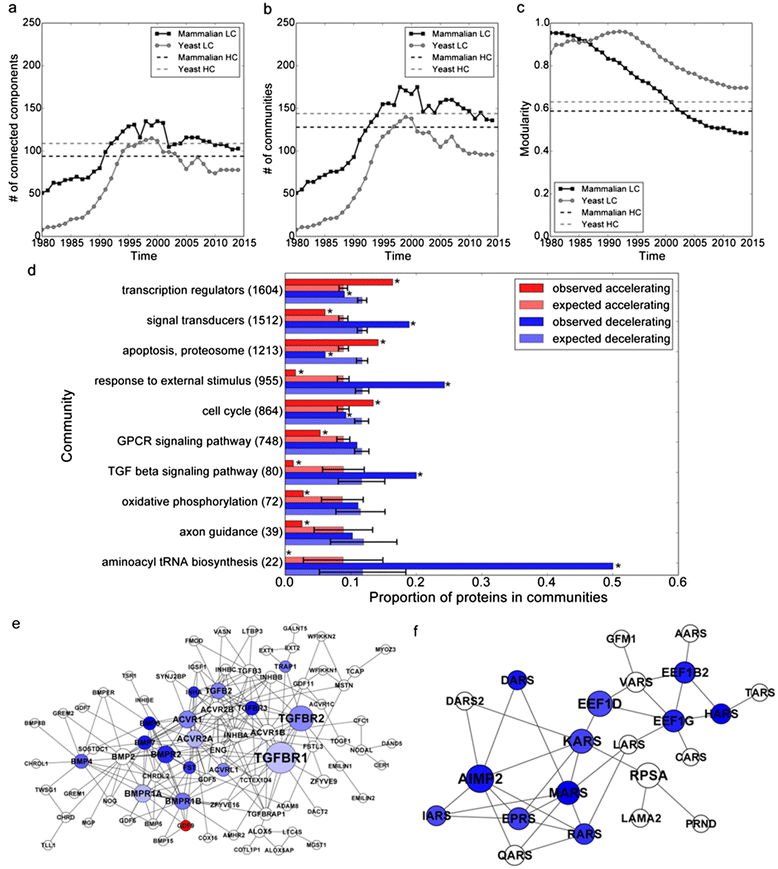
References
-
- Cordeddu V, Di Schiavi E, Pennacchio LA, Ma’ayan A, Sarkozy A, Fodale V, Cecchetti S, Cardinale A, Martin J, Schackwitz W. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat Genet. 2009;41(9):1022–6. doi: 10.1038/ng.425. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

