Loss of FERULATE 5-HYDROXYLASE Leads to Mediator-Dependent Inhibition of Soluble Phenylpropanoid Biosynthesis in Arabidopsis
- PMID: 26048881
- PMCID: PMC4634044
- DOI: 10.1104/pp.15.00294
Loss of FERULATE 5-HYDROXYLASE Leads to Mediator-Dependent Inhibition of Soluble Phenylpropanoid Biosynthesis in Arabidopsis
Abstract
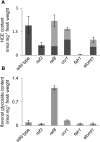
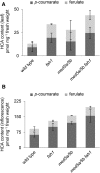
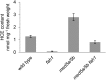
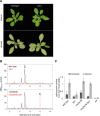
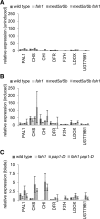
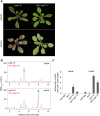
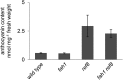
Phenylpropanoids are phenylalanine-derived specialized metabolites and include important structural components of plant cell walls, such as lignin and hydroxycinnamic acids, as well as ultraviolet and visible light-absorbing pigments, such as hydroxycinnamate esters (HCEs) and anthocyanins. Previous work has revealed a remarkable degree of plasticity in HCE biosynthesis, such that most Arabidopsis (Arabidopsis thaliana) mutants with blockages in the pathway simply redirect carbon flux to atypical HCEs. In contrast, the ferulic acid hydroxylase1 (fah1) mutant accumulates greatly reduced levels of HCEs, suggesting that phenylpropanoid biosynthesis may be repressed in response to the loss of FERULATE 5-HYDROXYLASE (F5H) activity. Here, we show that in fah1 mutant plants, the activity of HCE biosynthetic enzymes is not limiting for HCE accumulation, nor is phenylpropanoid flux diverted to the synthesis of cell wall components or flavonol glycosides. We further show that anthocyanin accumulation is also repressed in fah1 mutants and that this repression is specific to tissues in which F5H is normally expressed. Finally, we show that repression of both HCE and anthocyanin biosynthesis in fah1 mutants is dependent on the MED5a/5b subunits of the transcriptional coregulatory complex Mediator, which are similarly required for the repression of lignin biosynthesis and the stunted growth of the phenylpropanoid pathway mutant reduced epidermal fluorescence8. Taken together, these observations show that the synthesis of HCEs and anthocyanins is actively repressed in a MEDIATOR-dependent manner in Arabidopsis fah1 mutants and support an emerging model in which MED5a/5b act as central players in the homeostatic repression of phenylpropanoid metabolism.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 - PubMed
-
- Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M, et al. (2014) Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509: 376–380 - PubMed
-
- Chang XF, Chandra R, Berleth T, Beatson RP (2008) Rapid, microscale, acetyl bromide-based method for high-throughput determination of lignin content in Arabidopsis thaliana. J Agric Food Chem 56: 6825–6834 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

