Replication fork progression during re-replication requires the DNA damage checkpoint and double-strand break repair
- PMID: 26051888
- PMCID: PMC4580973
- DOI: 10.1016/j.cub.2015.04.058
Replication fork progression during re-replication requires the DNA damage checkpoint and double-strand break repair
Abstract
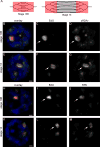
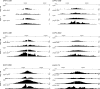
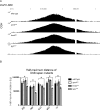
Replication origins are under tight regulation to ensure activation occurs only once per cell cycle [1, 2]. Origin re-firing in a single S phase leads to the generation of DNA double-strand breaks (DSBs) and activation of the DNA damage checkpoint [2-7]. If the checkpoint is blocked, cells enter mitosis with partially re-replicated DNA that generates chromosome breaks and fusions [5]. These types of chromosomal aberrations are common in numerous human cancers, suggesting that re-replication events contribute to cancer progression. It was proposed that fork instability and DSBs formed during re-replication are the result of head-to-tail collisions and collapse of adjacent replication forks [3]. However, previously studied systems lack the resolution to determine whether the observed DSBs are generated at sites of fork collisions. Here, we utilize the Drosophila ovarian follicle cells, which exhibit re-replication under precise developmental control [8-10], to model the consequences of re-replication at actively elongating forks. Re-replication occurs from specific replication origins at six genomic loci, termed Drosophila amplicons in follicle cells (DAFCs) [10-12]. Precise developmental timing of DAFC origin firing permits identification of forks at defined points after origin initiation [13, 14]. Here, we show that DAFC re-replication causes fork instability and generates DSBs at sites of potential fork collisions. Immunofluorescence and ChIP-seq demonstrate the DSB marker γH2Av is enriched at elongating forks. Fork progression is reduced in the absence of DNA damage checkpoint components and nonhomologous end-joining (NHEJ), but not homologous recombination. NHEJ appears to continually repair forks during re-replication to maintain elongation.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

References
-
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

