Site-specific processing of Ras and Rap1 Switch I by a MARTX toxin effector domain
- PMID: 26051945
- PMCID: PMC4468845
- DOI: 10.1038/ncomms8396
Site-specific processing of Ras and Rap1 Switch I by a MARTX toxin effector domain
Abstract
Ras (Rat sarcoma) protein is a central regulator of cell growth and proliferation. Mutations in the RAS gene are known to occur in human cancers and have been shown to contribute to carcinogenesis. In this study, we show that the multifunctional-autoprocessing repeats-in-toxin (MARTX) toxin-effector domain DUF5(Vv) from Vibrio vulnificus to be a site-specific endopeptidase that cleaves within the Switch 1 region of Ras and Rap1. DUF5(Vv) processing of Ras, which occurs both biochemically and in mammalian cell culture, inactivates ERK1/2, thereby inhibiting cell proliferation. The ability to cleave Ras and Rap1 is shared by DUF5(Vv) homologues found in other bacteria. In addition, DUF5(Vv )can cleave all Ras isoforms and KRas with mutations commonly implicated in malignancies. Therefore, we speculate that this new family of Ras/Rap1-specific endopeptidases (RRSPs) has potential to inactivate both wild-type and mutant Ras proteins expressed in malignancies.
Conflict of interest statement
I.A., M.B. and K.J.F.S. have submitted a provisional patent application covering this research. The remaining authors declares no competing financial interest.
Figures
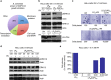
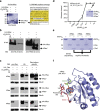


Comment in
-
A bacterial toxin that cleaves Ras oncoprotein.Oncotarget. 2015 Aug 7;6(22):18742-3. doi: 10.18632/oncotarget.5113. Oncotarget. 2015. PMID: 26300052 Free PMC article. No abstract available.
References
-
- Santos E. et al.. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science 223, 661–664 (1984). - PubMed
-
- Malumbres M. & Barbacid M. RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3, 459–465 (2003). - PubMed
-
- Young A., Lou D. & McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 3, 112–123 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01CA168292/CA/NCI NIH HHS/United States
- R01 AI092825/AI/NIAID NIH HHS/United States
- R01 CA152799/CA/NCI NIH HHS/United States
- R37 AI092825/AI/NIAID NIH HHS/United States
- R01CA152601/CA/NCI NIH HHS/United States
- R01AI051490/AI/NIAID NIH HHS/United States
- R01 CA168292/CA/NCI NIH HHS/United States
- R01 CA152601/CA/NCI NIH HHS/United States
- R01CA152799/CA/NCI NIH HHS/United States
- R01 AI051490/AI/NIAID NIH HHS/United States
- R01 AI098369/AI/NIAID NIH HHS/United States
- R01AI098369/AI/NIAID NIH HHS/United States
- R01AI092825/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

