The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling
- PMID: 26053220
- PMCID: PMC4630029
- DOI: 10.1038/ncb3179
The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling
Abstract
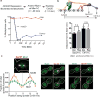
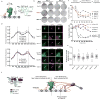
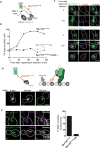
The spindle assembly checkpoint (SAC) is a unique signalling mechanism that responds to the state of attachment of the kinetochore to spindle microtubules. SAC signalling is activated by unattached kinetochores, and it is silenced after these kinetochores form end-on microtubule attachments. Although the biochemical cascade of SAC signalling is well understood, how kinetochore-microtubule attachment disrupts it remained unknown. Here we show that, in budding yeast, end-on microtubule attachment to the kinetochore physically separates the Mps1 kinase, which probably binds to the calponin homology domain of Ndc80, from the kinetochore substrate of Mps1, Spc105 (KNL1 orthologue). This attachment-mediated separation disrupts the phosphorylation of Spc105, and enables SAC silencing. Additionally, the Dam1 complex may act as a barrier that shields Spc105 from Mps1. Together these data suggest that the protein architecture of the kinetochore encodes a mechanical switch. End-on microtubule attachment to the kinetochore turns this switch off to silence the SAC.
Figures




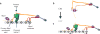
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

