Diversity of lactase persistence in African milk drinkers
- PMID: 26054462
- PMCID: PMC4495257
- DOI: 10.1007/s00439-015-1573-2
Diversity of lactase persistence in African milk drinkers
Abstract
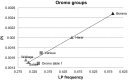
The genetic trait of lactase persistence is attributable to allelic variants in an enhancer region upstream of the lactase gene, LCT. To date, five different functional alleles, -13910*T, -13907*G, -13915*G, -14009*G and -14010*C, have been identified. The co-occurrence of several of these alleles in Ethiopian lactose digesters leads to a pattern of sequence diversity characteristic of a 'soft selective sweep'. Here we hypothesise that throughout Africa, where multiple functional alleles co-exist, the enhancer diversity will be greater in groups who are traditional milk drinkers than in non-milk drinkers, as the result of this sort of parallel selection. Samples from 23 distinct groups from 10 different countries were examined. Each group was classified 'Yes 'or 'No' for milk-drinking, and ethnicity, language spoken and geographic location were recorded. Predicted lactase persistence frequency and enhancer diversity were, as hypothesised, higher in the milk drinkers than the non-milk-drinkers, but this was almost entirely accounted for by the Afro-Asiatic language speaking peoples of east Africa. The other groups, including the 'Nilo-Saharan language speaking' milk-drinkers, show lower frequencies of LP and lower diversity, and there was a north-east to south-west decline in overall diversity. Amongst the Afro-Asiatic (Cushitic) language speaking Oromo, however, the geographic cline was not evident and the southern pastoralist Borana showed much higher LP frequency and enhancer diversity than the other groups. Together these results reflect the effects of parallel selection, the stochastic processes of the occurrence and spread of the mutations, and time depth of milk drinking tradition.
Figures



References
-
- Arnold J, Diop M, Kodjovi M, Rozier J. Lactose intolerance in adults in Senegal. C R Seances Soc Biol Fil. 1980;174:983–992. - PubMed
-
- Baddeley A, Turner R. Spatstat: an R package for analyzing spatial point patterns. J Stat Softw. 2005;12:1–42.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

