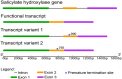
Functional characterization of salicylate hydroxylase from the fungal endophyte Epichloë festucae
- PMID: 26055188
- PMCID: PMC4460724
- DOI: 10.1038/srep10939
Functional characterization of salicylate hydroxylase from the fungal endophyte Epichloë festucae
Abstract
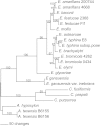
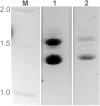
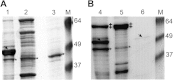
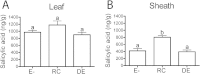
Epichloë spp. are symbiotic fungal endophytes of many cool season grasses. The presence of the fungal endophytes often confers insect, drought, and disease tolerance to the host grasses. The presence of the fungal endophytes within the host plants does not elicit host defense responses. The molecular basis for this phenomenon is not known. Epichloë festucae, the endophyte of Festuca rubra, expresses a salicylate hydroxylase similar to NahG from the bacterium Pseudomonas putida. Few fungal salicylate hydroxylase enzymes have been reported. The in planta expression of an endophyte salicylate hydroxylase raised the possibility that degradation of plant-produced salicylic acid is a factor in the mechanism of how the endophyte avoids eliciting host plant defenses. Here we report the characterization of the E. festucae salicylate hydroxylase, designated Efe-shyA. Although the fungal enzyme has the expected activity, based on salicylic acid levels in endophyte-free and endophyte-infected plants it is unlikely that expression of the endophyte salicylate hydroxylase is a factor in the lack of a host defense response to the presence of the fungal endophyte.
Figures



References
-
- Leuchtmann A., Bacon C. W., Schardl C. L., White J. F. Jr. & Tadych M. Nomenclatural realignment of Neotyphodium species with genus Epichloë. Mycologia 106, 202–215 (2014). - PubMed
-
- Kuldau G. & Bacon C. Clavicipitaceous endophytes: their ability to enhance resistance of grasses to multiple stresses. Biol. Control 46, 57–71 (2008).
-
- Schardl C. L., Scott B., Florea S. & Zhang D. Epichloë endophytes: clavicipitaceous symbionts of grasses. In Plant Relationships, 2nd Edition, The Mycota V (ed Deising H. ) 275–306 (Springer-Verlag, Berlin Heidelberg, 2009).
-
- Clarke B. B. et al.. Endophyte-mediated suppression of dollar spot disease in fine fescues. Plant Dis. 90, 994–998 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

