Novel players in coeliac disease pathogenesis: role of the gut microbiota
- PMID: 26055247
- PMCID: PMC5102016
- DOI: 10.1038/nrgastro.2015.90
Novel players in coeliac disease pathogenesis: role of the gut microbiota
Abstract
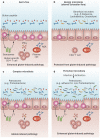
Several studies point towards alteration in gut microbiota composition and function in coeliac disease, some of which can precede the onset of disease and/or persist when patients are on a gluten-free diet. Evidence also exists that the gut microbiota might promote or reduce coeliac-disease-associated immunopathology. However, additional studies are required in humans and in mice (using gnotobiotic technology) to determine cause-effect relationships and to identify agents for modulating the gut microbiota as a therapeutic or preventative approach for coeliac disease. In this Review, we summarize the current evidence for altered gut microbiota composition in coeliac disease and discuss how the interplay between host genetics, environmental factors and the intestinal microbiota might contribute to its pathogenesis. Moreover, we highlight the importance of utilizing animal models and long-term clinical studies to gain insight into the mechanisms through which host-microbial interactions can influence host responses to gluten.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

