A Region of Bdp1 Necessary for Transcription Initiation That Is Located within the RNA Polymerase III Active Site Cleft
- PMID: 26055328
- PMCID: PMC4508317
- DOI: 10.1128/MCB.00263-15
A Region of Bdp1 Necessary for Transcription Initiation That Is Located within the RNA Polymerase III Active Site Cleft
Abstract
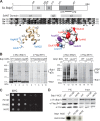
The RNA polymerase III (Pol III)-specific transcription factor Bdp1 is crucial to Pol III recruitment and promoter opening in transcription initiation, yet structural information is sparse. To examine its protein-binding targets within the preinitiation complex at the residue level, photoreactive amino acids were introduced into Saccharomyces cerevisiae Bdp1. Mutations within the highly conserved SANT domain cross-linked to the transcription factor IIB (TFIIB)-related transcription factor Brf1, consistent with the findings of previous studies. In addition, we identified an essential N-terminal region that cross-linked with the Pol III catalytic subunit C128 as well as Brf1. Closer examination revealed that this region interacted with the C128 N-terminal region, the N-terminal half of Brf1, and the C-terminal domain of the C37 subunit, together positioning this region within the active site cleft of the preinitiation complex. With our functional data, our analyses identified an essential region of Bdp1 that is positioned within the active site cleft of Pol III and necessary for transcription initiation.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




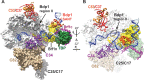
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
