Insights into the Recruitment of Class IIa Histone Deacetylases (HDACs) to the SMRT/NCoR Transcriptional Repression Complex
- PMID: 26055705
- PMCID: PMC4505066
- DOI: 10.1074/jbc.M115.661058
Insights into the Recruitment of Class IIa Histone Deacetylases (HDACs) to the SMRT/NCoR Transcriptional Repression Complex
Abstract
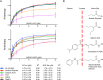
Class IIa histone deacetylases repress transcription of target genes. However, their mechanism of action is poorly understood because they exhibit very low levels of deacetylase activity. The class IIa HDACs are associated with the SMRT/NCoR repression complexes and this may, at least in part, account for their repressive activity. However, the molecular mechanism of recruitment to co-repressor proteins has yet to be established. Here we show that a repeated peptide motif present in both SMRT and NCoR is sufficient to mediate specific interaction, with micromolar affinity, with all the class IIa HDACs (HDACs 4, 5, 7, and 9). Mutations in the consensus motif abrogate binding. Mutational analysis of HDAC4 suggests that the peptide interacts in the vicinity of the active site of the enzyme and requires the "closed" conformation of the zinc-binding loop on the surface of the enzyme. Together these findings represent the first insights into the molecular mechanism of recruitment of class IIa HDACs to the SMRT/NCoR repression complexes.
Keywords: epigenetics; histone acetylation; histone deacetylase 4 (HDAC4); protein-protein interaction; transcription corepressor.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 - PubMed
-
- Hildmann C., Riester D., Schwienhorst A. (2007) Histone deacetylases: an important class of cellular regulators with a variety of functions. Appl. Microbiol. Biotechnol. 75, 487–497 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

