Underexpression of LKB1 tumor suppressor is associated with enhanced Wnt signaling and malignant characteristics of human intrahepatic cholangiocarcinoma
- PMID: 26056085
- PMCID: PMC4662463
- DOI: 10.18632/oncotarget.4305
Underexpression of LKB1 tumor suppressor is associated with enhanced Wnt signaling and malignant characteristics of human intrahepatic cholangiocarcinoma
Abstract
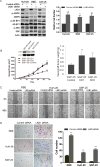
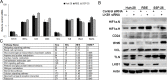
Intrahepatic cholangiocarcinoma (ICC) is a rare and highly aggressive malignancy. In this study, we identified the presence of gene deletion and missense mutation leading to inactivation or underexpression of liver kinase B1 (LKB1) tumor suppressor and excluded the involvement of LKB1 gene hypermethylation in ICC tissues. Immunohistochemical analysis showed that LKB1 was underexpressed in a portion of 326 ICC tissues compared to their adjacent normal tissues. By statistical analysis underexpression of LKB1 in ICC tissues significantly correlated with poor survival and malignant disease characteristics in ICC patients. Moreover, we showed that knockdown of LKB1 significantly enhanced growth, migration, and invasion of three LKB1-competent ICC cell lines. Global transcriptional profiling analysis identified multiple malignancy-promoting genes, such as HIF-1α, CD24, Talin1, Vinculin, Wnt5, and signaling pathways including Hedgehog, Wnt/β-catenin, and cell adhesion as novel targets of LKB1 underexpression in ICC cells. Furthermore, knockdown of LKB1 gene expression dramatically enhanced Wnt/β-catenin signaling in ICC cells, while an inverse correlation between LKB1 and nuclear β-catenin was observed in ICC tissues. Our findings suggest a novel mechanism for ICC carcinogenesis in which LKB1 underexpression enhances multiple signaling pathways including Wnt/β-catenin to promote disease progression.
Keywords: Wnt/β-catenin; global transcriptional profiling; intrahepatic cholangiocarcinoma (ICC); liver kinase B1 (LKB1); recurrence and metastasis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bridgewater J, Galle PR, Khan SA, Llovet JM, Park JW, Patel T, Pawlik TM, et al. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J Hepatol. 2014;60:1268–1289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

