Host lysozyme-mediated lysis of Lactococcus lactis facilitates delivery of colitis-attenuating superoxide dismutase to inflamed colons
- PMID: 26056274
- PMCID: PMC4485081
- DOI: 10.1073/pnas.1501897112
Host lysozyme-mediated lysis of Lactococcus lactis facilitates delivery of colitis-attenuating superoxide dismutase to inflamed colons
Abstract
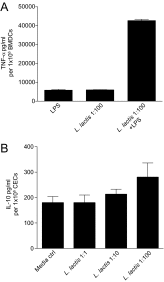
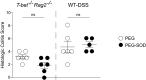
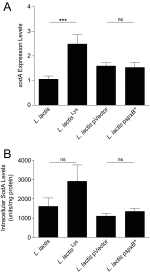
Beneficial microbes that target molecules and pathways, such as oxidative stress, which can negatively affect both host and microbiota, may hold promise as an inflammatory bowel disease therapy. Prior work showed that a five-strain fermented milk product (FMP) improved colitis in T-bet(-/-) Rag2(-/-) mice. By varying the number of strains used in the FMP, we found that Lactococcus lactis I-1631 was sufficient to ameliorate colitis. Using comparative genomic analyses, we identified genes unique to L. lactis I-1631 involved in oxygen respiration. Respiration of oxygen results in reactive oxygen species (ROS) generation. Also, ROS are produced at high levels during intestinal inflammation and cause tissue damage. L. lactis I-1631 possesses genes encoding enzymes that detoxify ROS, such as superoxide dismutase (SodA). Thus, we hypothesized that lactococcal SodA played a role in attenuating colitis. Inactivation of the sodA gene abolished L. lactis I-1631's beneficial effect in the T-bet(-/-) Rag2(-/-) model. Similar effects were obtained in two additional colonic inflammation models, Il10(-/-) mice and dextran sulfate sodium-treated mice. Efforts to understand how a lipophobic superoxide anion (O2 (-)) can be detoxified by cytoplasmic lactoccocal SodA led to the finding that host antimicrobial-mediated lysis is a prerequisite for SodA release and SodA's extracytoplasmic O2 (-) scavenging. L. lactis I-1631 may represent a promising vehicle to deliver antioxidant, colitis-attenuating SodA to the inflamed intestinal mucosa, and host antimicrobials may play a critical role in mediating SodA's bioaccessibility.
Keywords: Lactococcus lactis; colitis; lysozyme; oxidative stress; probiotics.
Conflict of interest statement
Conflict of interest statement: P.V., G.Q., P.G., C.B., M.D., and J.v.H.V. are employees of and hold equity in Danone Nutricia Research. This study was supported by a grant from Danone Nutricia Research, R01CA154426 (National Cancer Institute), K08AI078942 (National Institute of Allergy and Infectious Diseases), a Burroughs Wellcome Career in Medical Sciences Award, and a Searle Scholars Award.
Figures




Comment in
-
IBD: Lactococcus lactis alleviates oxidative stress and colitis in mice.Nat Rev Gastroenterol Hepatol. 2015 Aug;12(8):429. doi: 10.1038/nrgastro.2015.109. Epub 2015 Jun 23. Nat Rev Gastroenterol Hepatol. 2015. PMID: 26100371 No abstract available.
References
-
- Uhlig HH, et al. 2014. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology 147(5):990–1007.e3.
-
- Packey CD, Sartor RB. Interplay of commensal and pathogenic bacteria, genetic mutations, and immunoregulatory defects in the pathogenesis of inflammatory bowel diseases. J Intern Med. 2008;263(6):597–606. - PubMed
-
- Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474(7351):298–306. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

